Genetic Variation Controlling Wrinkled Seed Phenotypes in Pisum: How Lucky Was Mendel?
- PMID: 28587311
- PMCID: PMC5486028
- DOI: 10.3390/ijms18061205
Genetic Variation Controlling Wrinkled Seed Phenotypes in Pisum: How Lucky Was Mendel?
Abstract
One of the traits studied by Mendel in pea (Pisum sativum L.) was the wrinkled-seeded phenotype, and the molecular basis for a mutation underlying this phenotype was discovered in the 1990s. Although the starch-branching enzyme gene mutation identified at the genetic locus r is most likely to be that in seeds available to Mendel in the mid-1800s, it has remained an open question as to whether or not additional natural mutations in this gene exist within Pisum germplasm collections. Here, we explore this question and show that all but two wrinkled-seeded variants in one such collection correspond to either the mutant allele described previously for the r locus or a mutation at a second genetic locus, rb, affecting the gene encoding the large subunit of Adenosine diphosphoglucose (ADP-glucose) pyrophosphorylase; the molecular basis for the rb mutation is described here. The genetic basis for the phenotype of one (JI 2110) of the two lines which are neither r nor rb has been studied in crosses with a round-seeded variant (JI 281); for which extensive genetic marker data were expected. In marked contrast to the trait studied by Mendel and the rb phenotype; the data suggest that the wrinkled-seeded phenotype in JI 2110 is maternally determined, controlled by two genetic loci, and the extent to which it is manifested is very sensitive to the environment. Metabolite analysis of the cotyledons of JI 2110 revealed a profile for sucrose and sucrose-derived compounds that was more similar to that of wild-type round-seeded, than that of wrinkled-seeded r, pea lines. However, the metabolite profile of the seed coat (testa) of JI 2110 was distinct from that of other round-seeded genotypes tested which, together with analysis of recombinant inbred progeny lines, suggests an explanation for the seed phenotype.
Keywords: genetic markers; myoinositol; pea germplasm; r and rb mutations; seed coat (testa) metabolites; seed phenotype; wrinkled seeds.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


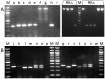
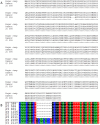
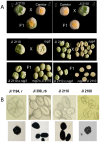

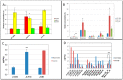
References
-
- Wang T.L., Bogracheva T.Y., Hedley C.L. Starch: As simple as A, B, C? J. Exp. Bot. 1998;49:481–502. doi: 10.1093/jxb/49.320.481. - DOI
-
- Mendel G. Versuche über pflanzen-hybriden. Verhandlungen des Naturforschenden Vereines in Brünn. 1866;4:3–47.
-
- Wang T.L., Hedley C.L. Seed development in peas: Knowing your three “r’s” (or four, or five) Seed Sci. Res. 1991;1:3–14. doi: 10.1017/S096025850000057X. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

