Proteomic analysis by iTRAQ-MRM of soybean resistance to Lamprosema Indicate
- PMID: 28587595
- PMCID: PMC5461738
- DOI: 10.1186/s12864-017-3825-0
Proteomic analysis by iTRAQ-MRM of soybean resistance to Lamprosema Indicate
Abstract
Background: Lamprosema indicate is a major leaf feeding insect pest to soybean, which has caused serious yield losses in central and southern China. To explore the defense mechanisms of soybean resistance to Lamprosema indicate, a highly resistant line (Gantai-2-2) and a highly susceptible line (Wan 82-178) were exposed to Lamprosema indicate larval feedings for 0 h and 48 h, and the differential proteomic analyses of these two lines were carried out.
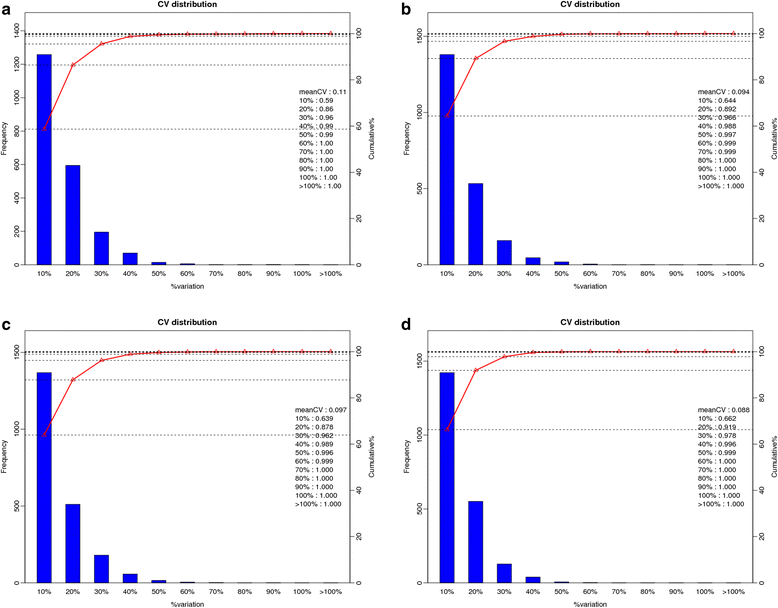
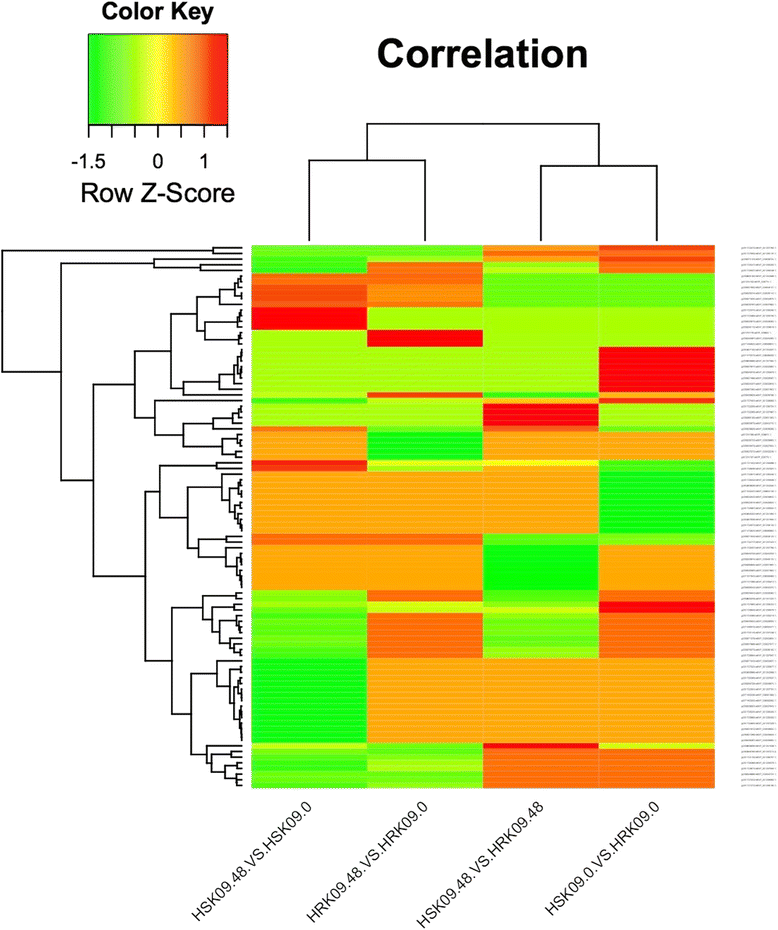
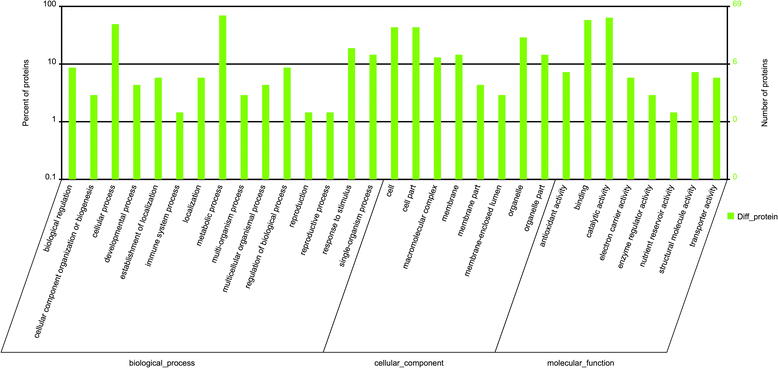
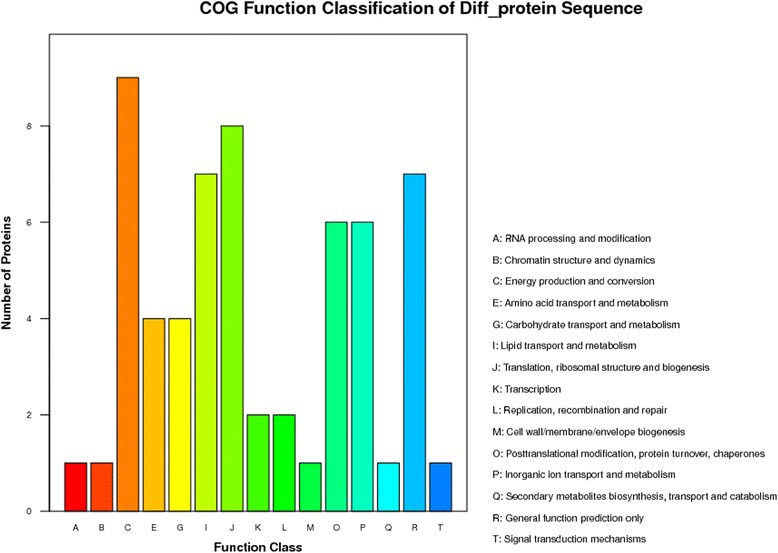
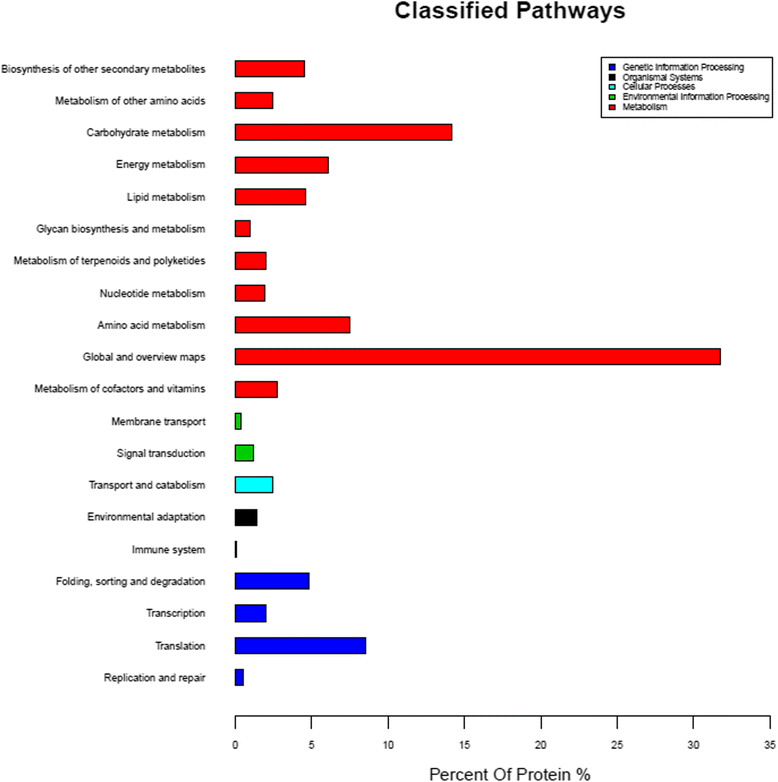
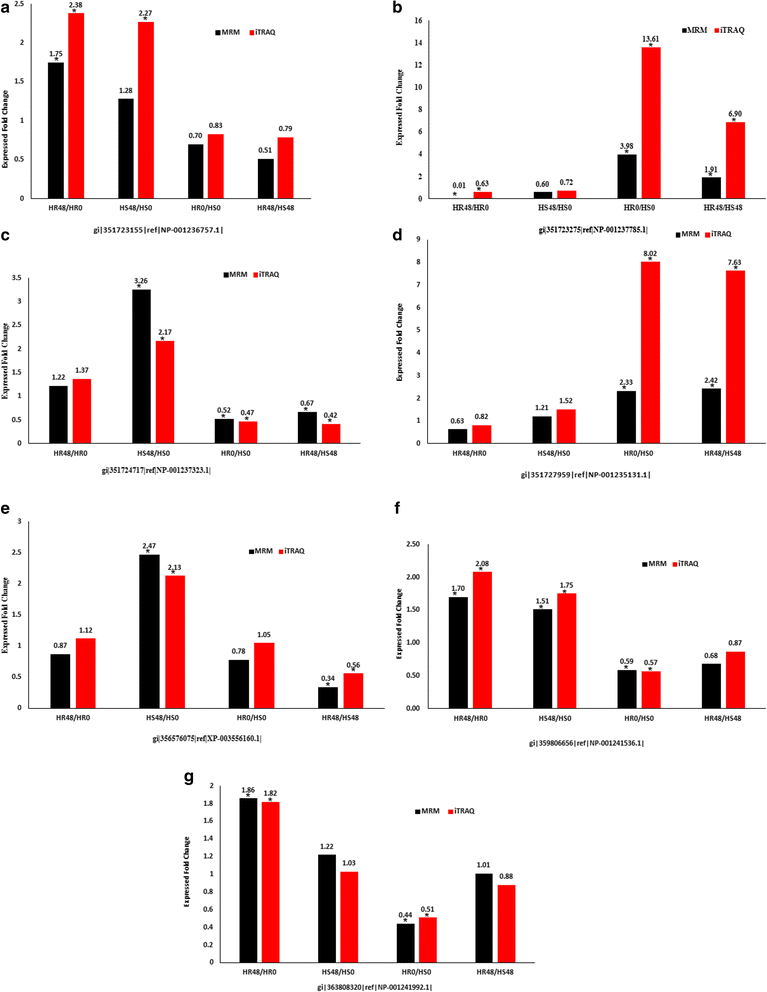
Results: The results showed that 31 differentially expressed proteins (DEPs) were identified in the Gantai-2-2 when comparing 48 h feeding with 0 h feeding, and 53 DEPs were identified in the Wan 82-178. 28 DEPs were identified when comparing Gantai-2-2 with Wan 82-178 at 0 h feeding. The bioinformatic analysis results showed that most of the DEPs were associated with ribosome, linoleic acid metabolism, flavonoid biosynthesis, phenylpropanoid biosynthesis, peroxisome, stilbenoid, diarylheptanoid and gingerol biosynthesis, glutathione metabolism, pant hormone signal transduction, and flavone and flavonol biosynthesis, as well as other resistance related metabolic pathways. The MRM analysis showed that the iTRAQ results were reliable.
Conclusions: According to the analysis of the DEPs results, the soybean defended or resisted the Lamprosema indicate damage by the induction of a synthesis of anti-digestive proteins which inhibit the growth and development of insects, reactive oxygen species scavenging, signaling pathways, secondary metabolites synthesis, and so on.
Keywords: Differentially expressed protein; Lamprosema indicate (Fabricius); MRM; Soybean; iTRAQ.
Figures
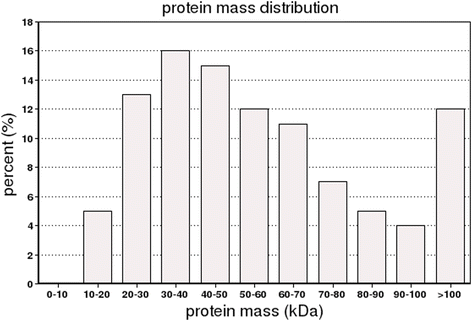
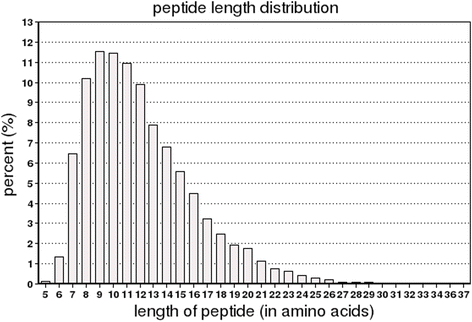
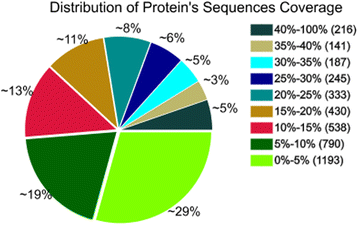
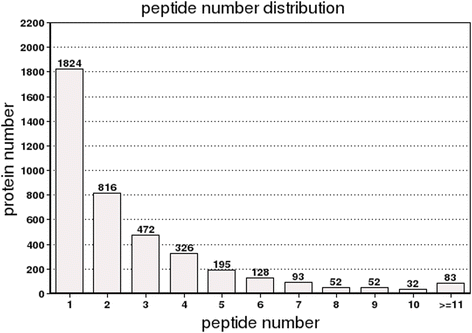
References
-
- Paiva NL. An introduction to the biosynthesis of chemicals used in plant-microbe communication. J Plant Growth Regul. 2000;19(2):131–143. - PubMed
-
- Sangha JS, Chen YH, Jatinder K, Wajahatullah K, Zainularifeen A, Mohammed SA, et al. Proteome analysis of rice (Oryza sativa L.) mutants reveals differentially induced proteins during Brown Planthopper (Nilaparvata lugens) infestation. Int J Mol Sci. 2013;14(2):3921–3945. doi: 10.3390/ijms14023921. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

