Improved detection of gene-microbe interactions in the mouse skin microbiota using high-resolution QTL mapping of 16S rRNA transcripts
- PMID: 28587635
- PMCID: PMC5461731
- DOI: 10.1186/s40168-017-0275-5
Improved detection of gene-microbe interactions in the mouse skin microbiota using high-resolution QTL mapping of 16S rRNA transcripts
Abstract
Background: Recent studies highlight the utility of quantitative trait locus (QTL) mapping for determining the contribution of host genetics to interindividual variation in the microbiota. We previously demonstrated that similar to the gut microbiota, abundances of bacterial taxa in the skin are significantly influenced by host genetic variation. In this study, we analyzed the skin microbiota of mice from the 15th generation of an advanced intercross line using a novel approach of extending bacterial trait mapping to both the 16S rRNA gene copy (DNA) and transcript (RNA) levels, which reflect relative bacterial cell number and activity, respectively.
Results: Remarkably, the combination of highly recombined individuals and 53,203 informative SNPs allowed the identification of genomic intervals as small as <0.1 megabases containing single genes. Furthermore, the inclusion of 16S rRNA transcript-level mapping dramatically increased the number of significant associations detected, with five versus 21 significant SNP-bacterial trait associations based on DNA- compared to RNA-level profiling, respectively. Importantly, the genomic intervals identified contain many genes involved in skin inflammation and cancer and are further supported by the bacterial traits they influence, which in some cases have known genotoxic or probiotic capabilities.
Conclusions: These results indicate that profiling based on the relative activity levels of bacterial community members greatly enhances the capability of detecting interactions between the host and its associated microbes. Finally, the identification of several genes involved in skin cancer suggests that similar to colon carcinogenesis, the resident microbiota may play a role in skin cancer susceptibility and its potential prevention and/or treatment.
Keywords: 16S rRNA transcript; QTL mapping; Skin cancer; Skin microbiota.
Figures
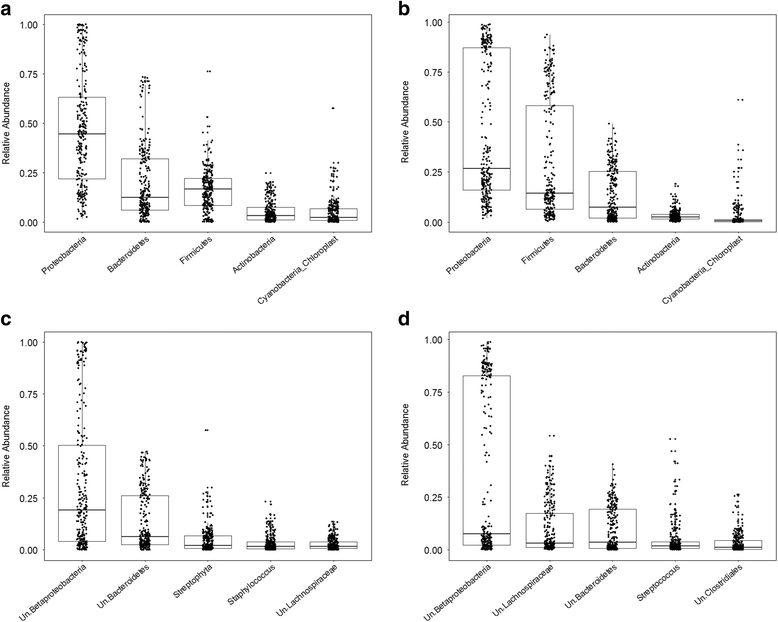

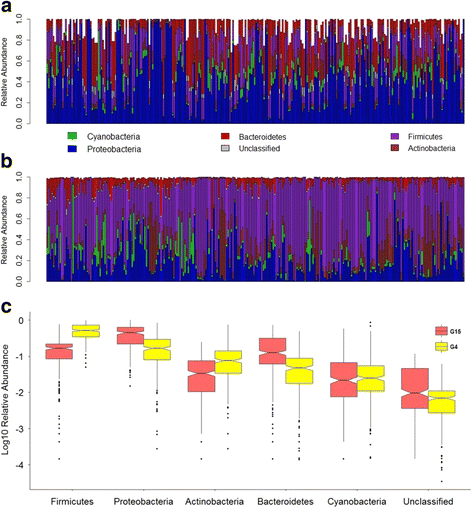
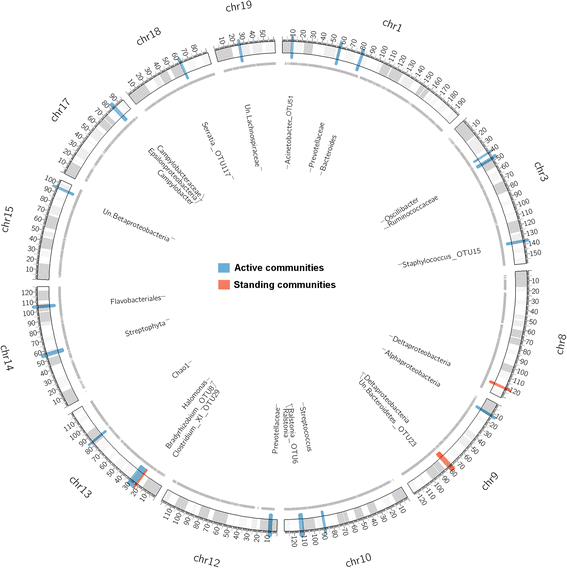

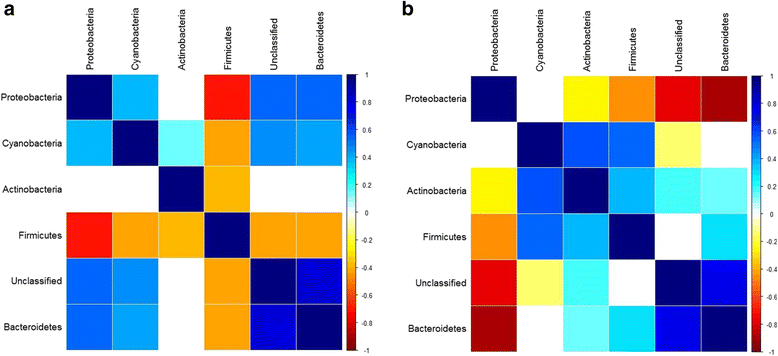
References
-
- McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PA, Bastiaansen JW, Wang X, Kachman SD, Auwerx J, Williams RW, Benson AK, Peterson DA, Ciobanu DC. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One. 2012;7(6):e39191. doi: 10.1371/journal.pone.0039191. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

