Comprehensive global genome dynamics of Chlamydia trachomatis show ancient diversification followed by contemporary mixing and recent lineage expansion
- PMID: 28588068
- PMCID: PMC5495073
- DOI: 10.1101/gr.212647.116
Comprehensive global genome dynamics of Chlamydia trachomatis show ancient diversification followed by contemporary mixing and recent lineage expansion
Abstract
Chlamydia trachomatis is the world's most prevalent bacterial sexually transmitted infection and leading infectious cause of blindness, yet it is one of the least understood human pathogens, in part due to the difficulties of in vitro culturing and the lack of available tools for genetic manipulation. Genome sequencing has reinvigorated this field, shedding light on the contemporary history of this pathogen. Here, we analyze 563 full genomes, 455 of which are novel, to show that the history of the species comprises two phases, and conclude that the currently circulating lineages are the result of evolution in different genomic ecotypes. Temporal analysis indicates these lineages have recently expanded in the space of thousands of years, rather than the millions of years as previously thought, a finding that dramatically changes our understanding of this pathogen's history. Finally, at a time when almost every pathogen is becoming increasingly resistant to antimicrobials, we show that there is no evidence of circulating genomic resistance in C. trachomatis.
© 2017 Hadfield et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

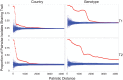
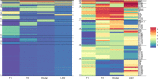
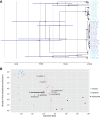

References
-
- Baker KS, Dallman TJ, Ashton PM, Day M, Hughes G, Crook PD, Gilbart VL, Zittermann S, Allen VG, Howden BP, et al. 2015. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect Dis 15: 913–921. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
