A Genoproteomic Approach to Detect Peptide Markers of Bacterial Respiratory Pathogens
- PMID: 28588123
- PMCID: PMC10863334
- DOI: 10.1373/clinchem.2016.269647
A Genoproteomic Approach to Detect Peptide Markers of Bacterial Respiratory Pathogens
Abstract
Background: Rapid identification of respiratory pathogens may facilitate targeted antimicrobial therapy. Direct identification of bacteria in bronchoalveolar lavage (BAL) by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry is confounded by interfering substances. We describe a method to identify unique peptide markers of 5 gram-negative bacteria by liquid chromatography-tandem mass spectrometry (LC-MS/MS) for direct pathogen identification in BAL.
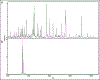
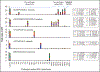
Methods: In silico translation and digestion were performed on 14-25 whole genomes representing strains of Acinetobacter baumannii, Moraxella catarrhalis, Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and Klebsiella pneumoniae. Peptides constituting theoretical core peptidomes in each were identified. Rapid tryptic digestion was performed; peptides were analyzed by LC-MS/MS and compared with the theoretical core peptidomes. High-confidence core peptides (false discovery rate <1%) were identified and analyzed with the lowest common ancestor search to yield potential species-specific peptide markers. The species specificity of each peptide was verified with protein BLAST. Further, 1 or 2 pathogens were serially diluted into pooled inflamed BAL, and a targeted LC-MS/MS assay was used to detect 25 peptides simultaneously.
Results: Five unique peptides with the highest abundance for each pathogen distinguished these pathogens with varied detection sensitivities. Peptide markers for A. baumannii and P. aeruginosa, when spiked simultaneously into inflamed BAL, were detected with as few as 3.6 (0.2) × 103 and 2.2 (0.6) × 103 colony-forming units, respectively, by targeted LC-MS/MS.
Conclusions: This proof-of-concept study shows the feasibility of identifying unique peptides in BAL for 5 gram-negative bacterial pathogens, and it may provide a novel approach for rapid direct identification of bacterial pathogens in BAL.
© 2017 American Association for Clinical Chemistry.
Conflict of interest statement
Figures

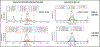
References
-
- Maschmeyer G, Carratala J, Buchheidt D, Hamprecht A, Heussel CP, Kahl C, et al. Diagnosis and antimicrobial therapy of lung infiltrates in febrile neutropenic patients (allogeneic SCT excluded): updated guidelines of the Infectious Diseases Working Party (AGIHO) of the German Society of Hematology and Medical Oncology (DGHO). Ann Oncol 2015;26:21–33. - PMC - PubMed
-
- Mikulska M, Del Bono V, Viscoli C. Bacterial infections in hematopoietic stem cell transplantation recipients. Curr Opin Hematol 2014;21:451–8. - PubMed
-
- Koncan R, Parisato M, Sakarikou C, Stringari G, Fontana C, Favuzzi V, et al. Direct identification of major gram-negative pathogens in respiratory specimens by respi-FISH® HAP Gram (−) panel, a beacon-based FISH methodology. Eur J Clin Microbiol Infect Dis 2015;34:2097–102. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

