Spatial Distribution and Ribosome-Binding Dynamics of EF-P in Live Escherichia coli
- PMID: 28588135
- PMCID: PMC5461404
- DOI: 10.1128/mBio.00300-17
Spatial Distribution and Ribosome-Binding Dynamics of EF-P in Live Escherichia coli
Abstract
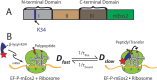
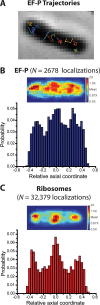
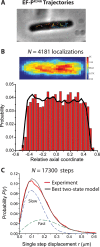
In vitro assays find that ribosomes form peptide bonds to proline (Pro) residues more slowly than to other residues. Ribosome profiling shows that stalling at Pro-Pro-X triplets is especially severe but is largely alleviated in Escherichia coli by the action of elongation factor EF-P. EF-P and its eukaryotic/archaeal homolog IF5A enhance the peptidyl transfer step of elongation. Here, a superresolution fluorescence localization and tracking study of EF-P-mEos2 in live E. coli provides the first in vivo information about the spatial distribution and on-off binding kinetics of EF-P. Fast imaging at 2 ms/frame helps to distinguish ribosome-bound (slowly diffusing) EF-P from free (rapidly diffusing) EF-P. Wild-type EF-P exhibits a three-peaked axial spatial distribution similar to that of ribosomes, indicating substantial binding. The mutant EF-PK34A exhibits a homogeneous distribution, indicating little or no binding. Some 30% of EF-P copies are bound to ribosomes at a given time. Two-state modeling and copy number estimates indicate that EF-P binds to 70S ribosomes during 25 to 100% of translation cycles. The timescale of the typical diffusive search by free EF-P for a ribosome-binding site is τfree ≈ 16 ms. The typical residence time of an EF-P on the ribosome is very short, τbound ≈ 7 ms. Evidently, EF-P binds to ribosomes during many or most elongation cycles, much more often than the frequency of Pro-Pro motifs. Emptying of the E site during part of the cycle is consistent with recent in vitro experiments indicating dissociation of the deacylated tRNA upon translocation.IMPORTANCE Ribosomes translate the codon sequence within mRNA into the corresponding sequence of amino acids within the nascent polypeptide chain, which in turn ultimately folds into functional protein. At each codon, bacterial ribosomes are assisted by two well-known elongation factors: EF-Tu, which aids binding of the correct aminoacyl-tRNA to the ribosome, and EF-G, which promotes tRNA translocation after formation of the new peptide bond. A third factor, EF-P, has been shown to alleviate ribosomal pausing at rare Pro-Pro motifs, which are translated very slowly without EF-P. Here, we use superresolution fluorescence imaging to study the spatial distribution and ribosome-binding dynamics of EF-P in live E. coli cells. We were surprised to learn that EF-P binds to and unbinds from translating ribosomes during at least 25% of all elongation events; it may bind during every elongation cycle.
Keywords: EF-P; binding dynamics; live E. coli; superresolution fluorescence.
Copyright © 2017 Mohapatra et al.
Figures


Comment in
-
Elongation Factor P Interactions with the Ribosome Are Independent of Pausing.mBio. 2017 Aug 1;8(4):e01056-17. doi: 10.1128/mBio.01056-17. mBio. 2017. PMID: 28765223 Free PMC article.
Similar articles
-
Simultaneous Binding of Multiple EF-Tu Copies to Translating Ribosomes in Live Escherichia coli.mBio. 2018 Jan 16;9(1):e02143-17. doi: 10.1128/mBio.02143-17. mBio. 2018. PMID: 29339430 Free PMC article.
-
Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.Mol Cell. 2017 Nov 2;68(3):515-527.e6. doi: 10.1016/j.molcel.2017.10.014. Mol Cell. 2017. PMID: 29100052
-
Substitution of Val20 by Gly in elongation factor Tu. Effects on the interaction with elongation factors Ts, aminoacyl-tRNA and ribosomes.Eur J Biochem. 1989 Nov 6;185(2):341-6. doi: 10.1111/j.1432-1033.1989.tb15121.x. Eur J Biochem. 1989. PMID: 2684669
-
The function of the translating ribosome: allosteric three-site model of elongation.Biochimie. 1991 Jul-Aug;73(7-8):1067-88. doi: 10.1016/0300-9084(91)90149-u. Biochimie. 1991. PMID: 1742351 Review.
-
Elongation Factor P and the Control of Translation Elongation.Annu Rev Microbiol. 2017 Sep 8;71:117-131. doi: 10.1146/annurev-micro-090816-093629. Epub 2017 May 22. Annu Rev Microbiol. 2017. PMID: 28886684 Review.
Cited by
-
Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.Nat Chem. 2023 Jan;15(1):143-153. doi: 10.1038/s41557-022-01073-1. Epub 2022 Oct 31. Nat Chem. 2023. PMID: 36316410 Free PMC article.
-
Near Saturation of Ribosomal L7/L12 Binding Sites with Ternary Complexes in Slowly Growing E. coli.J Mol Biol. 2019 May 31;431(12):2343-2353. doi: 10.1016/j.jmb.2019.04.037. Epub 2019 Apr 30. J Mol Biol. 2019. PMID: 31051175 Free PMC article.
-
Gut Microbiota in Untreated Diffuse Large B Cell Lymphoma Patients.Front Microbiol. 2021 Apr 13;12:646361. doi: 10.3389/fmicb.2021.646361. eCollection 2021. Front Microbiol. 2021. PMID: 33927704 Free PMC article.
-
Elongation factor P is required to maintain proteome homeostasis at high growth rate.Proc Natl Acad Sci U S A. 2018 Oct 23;115(43):11072-11077. doi: 10.1073/pnas.1812025115. Epub 2018 Oct 8. Proc Natl Acad Sci U S A. 2018. PMID: 30297417 Free PMC article.
-
Coupled Transcription-Translation in Prokaryotes: An Old Couple With New Surprises.Front Microbiol. 2021 Jan 21;11:624830. doi: 10.3389/fmicb.2020.624830. eCollection 2020. Front Microbiol. 2021. PMID: 33552035 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
