PCDH18 is frequently inactivated by promoter methylation in colorectal cancer
- PMID: 28588296
- PMCID: PMC5460281
- DOI: 10.1038/s41598-017-03133-w
PCDH18 is frequently inactivated by promoter methylation in colorectal cancer
Abstract
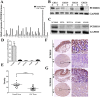
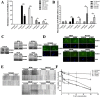
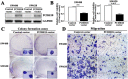
Protocadherin18 (PCDH18) was found to be preferentially methylated and inactivated in colorectal cancer (CRC) using bioinformatics tools. However, its biologic role in tumorgenesis remains unclear. Herein, we aimed to elucidate its epigenetic regulation and biological functions in CRC. The methylation status of PCDH18 was significant higher in CRC tissues than in adjacent non-tumor tissues (median, 15.17% vs. median, 0.4438%). Expression level of PCDH18 was significantly lower in primary CRCs than in nonmalignant tissues. Importantly, methylation status of PCDH18 in cell-free DNA of CRC patients was also significantly higher than in healthy subjects. PCDH18 was readily expressed in NCM460 cells, but downregulated in 100% (4/4) of CRC cell lines by promoter methylation, despite its expression could be restored through demethylation treatment. Overexpression of PCDH18 suppressed CRC cell viability, colony formation and migration. Meanwhile, the depletion of PCDH18 by siRNA in NCM460 cells enhanced the colonogenicity and migration ability and promoted β-catenin nuclear accumulation, whereas it inhibited cell cycle arrest. These effects were associated with upregulation of phospho-GSK-3β and cyclin D1, and downregulation of caspase3 and p21. Our results suggested that PCDH18 was a putative tumor suppressor with epigenetic silencing in CRC and a potential biomarker for CRC diagnosis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Zinc-finger protein 545 is inactivated due to promoter methylation and functions as a tumor suppressor through the Wnt/β-catenin, PI3K/AKT and MAPK/ERK signaling pathways in colorectal cancer.Int J Oncol. 2017 Sep;51(3):801-811. doi: 10.3892/ijo.2017.4064. Epub 2017 Jul 4. Int J Oncol. 2017. PMID: 28677721 Free PMC article.
-
LncRNA BDNF-AS suppresses colorectal cancer cell proliferation and migration by epigenetically repressing GSK-3β expression.Cell Biochem Funct. 2019 Jul;37(5):340-347. doi: 10.1002/cbf.3403. Epub 2019 May 6. Cell Biochem Funct. 2019. PMID: 31062382
-
Promoter hypermethylation of membrane type 3 matrix metalloproteinase is associated with cell migration in colorectal adenocarcinoma.Cancer Genet. 2015 May;208(5):261-70. doi: 10.1016/j.cancergen.2015.04.009. Epub 2015 May 1. Cancer Genet. 2015. PMID: 26002729
-
[Analysis of DNA methylation alterations in cellfree DNA fraction during colorectal cancer development].Magy Onkol. 2020 Mar 17;64(1):70-72. Epub 2019 Nov 21. Magy Onkol. 2020. PMID: 32181765 Review. Hungarian.
-
Epigenetic insights in the diagnosis, prognosis, and treatment selection in CRC, an updated review.Mol Biol Rep. 2022 Oct;49(10):10013-10022. doi: 10.1007/s11033-022-07569-w. Epub 2022 Jun 21. Mol Biol Rep. 2022. PMID: 35727475 Review.
Cited by
-
Spaceflight alters protein levels and gene expression associated with stress response and metabolic characteristics in human cardiac spheroids.Biomaterials. 2025 Jun;317:123080. doi: 10.1016/j.biomaterials.2024.123080. Epub 2025 Jan 6. Biomaterials. 2025. PMID: 39809079
-
JAM3 functions as a novel tumor suppressor and is inactivated by DNA methylation in colorectal cancer.Cancer Manag Res. 2019 Mar 27;11:2457-2470. doi: 10.2147/CMAR.S189937. eCollection 2019. Cancer Manag Res. 2019. PMID: 30988641 Free PMC article.
-
Mesenchyme-specific loss of Dot1L histone methyltransferase leads to skeletal dysplasia phenotype in mice.Bone. 2021 Jan;142:115677. doi: 10.1016/j.bone.2020.115677. Epub 2020 Oct 3. Bone. 2021. PMID: 33022452 Free PMC article.
-
Endothelial PAS domain protein 1 gene hypomethylation is associated with colorectal cancer in Han Chinese.Exp Ther Med. 2018 Dec;16(6):4983-4990. doi: 10.3892/etm.2018.6856. Epub 2018 Oct 12. Exp Ther Med. 2018. PMID: 30542453 Free PMC article.
-
Protocadherins at the Crossroad of Signaling Pathways.Front Mol Neurosci. 2020 Jun 30;13:117. doi: 10.3389/fnmol.2020.00117. eCollection 2020. Front Mol Neurosci. 2020. PMID: 32694982 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

