Intragenic CNTNAP2 Deletions: A Bridge Too Far?
- PMID: 28588433
- PMCID: PMC5448439
- DOI: 10.1159/000456021
Intragenic CNTNAP2 Deletions: A Bridge Too Far?
Abstract
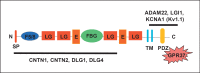
Intragenic deletions of the contactin-associated protein-like 2 gene (CNTNAP2) have been found in patients with Gilles de la Tourette syndrome, intellectual disability (ID), obsessive compulsive disorder, cortical dysplasia-focal epilepsy syndrome, autism, schizophrenia, Pitt-Hopkins syndrome, stuttering, and attention deficit hyperactivity disorder. A variety of molecular mechanisms, such as loss of transcription factor binding sites and perturbation of penetrance and expressivity, have been proposed to account for the phenotypic variability resulting from CNTNAP2 mutations. Deletions of both CNTNAP2 alleles produced truncated proteins lacking the transmembrane or some of the extracellular domains, or no protein at all. This observation can be extended to heterozygous intragenic deletions by assuming that such deletion-containing alleles lead to expression of a Caspr2 protein lacking one or several extracellular domains. Such altered forms of Capr2 proteins will lack the ability to bridge the intercellular space between neurons by binding to partners, such as CNTN1, CNTN2, DLG1, and DLG4. This presumed effect of intragenic deletions of CNTNAP2, and possibly other genes involved in connecting neuronal cells, represents a molecular basis for the postulated neuronal hypoconnectivity in autism and probably other neurodevelopmental disorders, including epilepsy, ID, language impairments and schizophrenia. Thus, CNTNAP2 may represent a paradigmatic case of a gene functioning as a node in a genetic and cellular network governing brain development and acquisition of higher cognitive functions.
Keywords: CNTNAP2; Intragenic deletions; Neurodevelopmental disorders.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

