Synthesis and Characterization of Novel BMI1 Inhibitors Targeting Cellular Self-Renewal in Hepatocellular Carcinoma
- PMID: 28589491
- PMCID: PMC7068645
- DOI: 10.1007/s11523-017-0501-x
Synthesis and Characterization of Novel BMI1 Inhibitors Targeting Cellular Self-Renewal in Hepatocellular Carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) represents one of the most lethal cancers worldwide due to therapy resistance and disease recurrence. Tumor relapse following treatment could be driven by the persistence of liver cancer stem-like cells (CSCs). The protein BMI1 is a member of the polycomb epigenetic factors governing cellular self-renewal, proliferation, and stemness maintenance. BMI1 expression also correlates with poor patient survival in various cancer types.
Objective: We aimed to elucidate the extent to which BMI1 can be used as a potential therapeutic target for CSC eradication in HCC.
Methods: We have recently participated in characterizing the first known pharmacological small molecule inhibitor of BMI1. Here, we synthesized a panel of novel BMI1 inhibitors and examined their ability to alter cellular growth and eliminate cancer progenitor/stem-like cells in HCC with different p53 backgrounds.
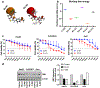
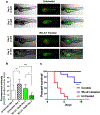
Results: Among various molecules examined, RU-A1 particularly downregulated BMI1 expression, impaired cell viability, reduced cell migration, and sensitized HCC cells to 5-fluorouracil (5-FU) in vitro. Notably, long-term analysis of HCC survival showed that, unlike chemotherapy, RU-A1 effectively reduced CSC content, even as monotherapy. BMI1 inhibition with RU-A1 diminished the number of stem-like cells in vitro more efficiently than the model compound C-209, as demonstrated by clonogenic assays and impairment of CSC marker expression. Furthermore, xenograft assays in zebrafish showed that RU-A1 abrogated tumor growth in vivo.
Conclusions: This study demonstrates the ability to identify agents with the propensity for targeting CSCs in HCC that could be explored as novel treatments in the clinical setting.
Conflict of interest statement
Figures




References
-
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–27. - PubMed
-
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362(9399):1907–17. - PubMed
-
- Yilmaz G, Akyol G, Cakir A, Ilhan M. Investigation of diagnostic utility and expression profiles of stem cell markers (CD133 and CD90) in hepatocellular carcinoma, small cell dysplasia, and cirrhosis. Pathol Res Pract. 2014;210(7):419–25. - PubMed
-
- Lee TK, Castilho A, Ma S, Ng IO. Liver cancer stem cells: implications for a new therapeutic target. Liver Int. 2009;29(7):955–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

