Comparative analyses of long non-coding RNA in lean and obese pig
- PMID: 28589911
- PMCID: PMC5522191
- DOI: 10.18632/oncotarget.18269
Comparative analyses of long non-coding RNA in lean and obese pig
Abstract
Objectives: Current studies have revealed that long non-coding RNA plays a crucial role in fat metabolism. However, the difference of lncRNA between lean (Duroc) and obese (Luchuan) pig remain undefined. Here, we investigated the expressional profile of lncRNA in these two pigs and discussed the relationship between lncRNA and fat deposition.
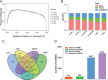
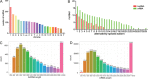
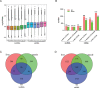
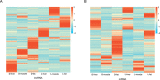
Materials and methods: The Chinese Luchuan pig has a dramatic differences in backfat thickness as compared with Duroc pig. In this study, 4868 lncRNA transcripts (including 3235 novel transcripts) were identified. We determined that patterns of differently expressed lncRNAs and mRNAs are strongly tissue-specific. The differentially expressed lncRNAs in adipose tissue have 794 potential target genes, which are involved in adipocytokine signaling pathways, the PI3k-Akt signaling pathway, and calcium signaling pathways. In addition, differentially expressed lncRNAs were located to 13 adipose-related quantitative trait loci which include 65 QTL_ID. Subsequently, lncRNA and mRNA in the same QTL_ID were analyzed and their co-expression in two QTL_ID were confirmed by qPCR.
Conclusions: Our study provides an insight into mechanism behind the fat metabolic differences between the two breeds and lays an important groundwork for further research regarding the regulatory role of lncRNA in obesity development.
Keywords: QTL; lncRNA; obesity; pig.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

