Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates
- PMID: 28590247
- PMCID: PMC5519329
- DOI: 10.7554/eLife.24133
Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates
Abstract
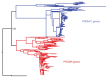
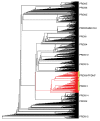
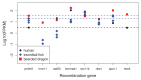
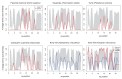
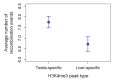
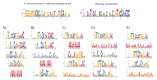
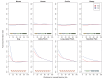
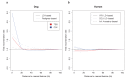
Studies of highly diverged species have revealed two mechanisms by which meiotic recombination is directed to the genome-through PRDM9 binding or by targeting promoter-like features-that lead to dramatically different evolutionary dynamics of hotspots. Here, we identify PRDM9 orthologs from genome and transcriptome data in 225 species. We find the complete PRDM9 ortholog across distantly related vertebrates but, despite this broad conservation, infer a minimum of six partial and three complete losses. Strikingly, taxa carrying the complete ortholog of PRDM9 are precisely those with rapid evolution of its predicted binding affinity, suggesting that all domains are necessary for directing recombination. Indeed, as we show, swordtail fish carrying only a partial but conserved ortholog share recombination properties with PRDM9 knock-outs.
Keywords: Hotspot; Meiosis; PRDM9; Recombination; Vertebrates; Xiphophorus; evolutionary biology; genomics.
Conflict of interest statement
MP: Reviewing editor,
The other authors declare that no competing interests exist.
Figures






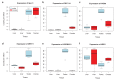
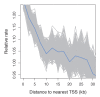
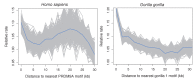
Comment in
-
Birth and death of a protein.Elife. 2017 Jul 20;6:e29502. doi: 10.7554/eLife.29502. Elife. 2017. PMID: 28726634 Free PMC article.
References
-
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, Ségurel L, Street T, Leffler EM, Bowden R, Aneas I, Broxholme J, Humburg P, Iqbal Z, Lunter G, Maller J, Hernandez RD, Melton C, Venkat A, Nobrega MA, Bontrop R, Myers S, Donnelly P, Przeworski M, McVean G. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

