Regulation of neural stem cell proliferation and differentiation by Kinesin family member 2a
- PMID: 28591194
- PMCID: PMC5462413
- DOI: 10.1371/journal.pone.0179047
Regulation of neural stem cell proliferation and differentiation by Kinesin family member 2a
Abstract
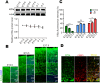
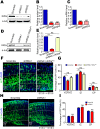
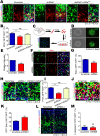
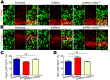
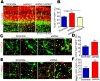
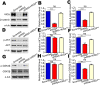
In the developing neocortex, cells in the ventricular/subventricular zone are largely multipotent neural stem cells and neural progenitor cells. These cells undergo self-renewal at the early stage of embryonic development to amplify the progenitor pool and subsequently differentiate into neurons. It is thus of considerable interest to investigate mechanisms controlling the switch from neural stem cells or neural progenitor cells to neurons. Here, we present evidence that Kif2a, a member of the Kinesin-13 family, plays a role in regulating the proliferation and differentiation of neural stem cells or neural progenitor cells at embryonic day 13.5. Silencing Kif2a by use of in utero electroporation of Kif2a shRNA reduced neural stem cells proliferation or self-renewal but increased neuronal differentiation. We further found that knockdown of Kif2a decreased the protein level of β-catenin, which is a critical molecule for neocortical neurogenesis. Together, these results reveal an important function of Kif2a in embryonic neocortical neurogenesis.
Conflict of interest statement
Figures
Similar articles
-
Hes5 regulates the transition timing of neurogenesis and gliogenesis in mammalian neocortical development.Development. 2017 Sep 1;144(17):3156-3167. doi: 10.1242/dev.147256. Development. 2017. PMID: 28851724
-
Hbp1 regulates the timing of neuronal differentiation during cortical development by controlling cell cycle progression.Development. 2015 Jul 1;142(13):2278-90. doi: 10.1242/dev.120477. Epub 2015 Jun 3. Development. 2015. PMID: 26041766
-
Hes1 overexpression leads to expansion of embryonic neural stem cell pool and stem cell reservoir in the postnatal brain.Development. 2021 Feb 17;148(4):dev189191. doi: 10.1242/dev.189191. Development. 2021. PMID: 33531431
-
Sonic hedgehog signaling coordinates the proliferation and differentiation of neural stem/progenitor cells by regulating cell cycle kinetics during development of the neocortex.Congenit Anom (Kyoto). 2012 Jun;52(2):72-7. doi: 10.1111/j.1741-4520.2012.00368.x. Congenit Anom (Kyoto). 2012. PMID: 22639991 Review.
-
Transcriptional Regulators and Human-Specific/Primate-Specific Genes in Neocortical Neurogenesis.Int J Mol Sci. 2020 Jun 29;21(13):4614. doi: 10.3390/ijms21134614. Int J Mol Sci. 2020. PMID: 32610533 Free PMC article. Review.
Cited by
-
KIF2A deficiency causes early-onset neurodegeneration.Proc Natl Acad Sci U S A. 2022 Nov 16;119(46):e2209714119. doi: 10.1073/pnas.2209714119. Epub 2022 Nov 7. Proc Natl Acad Sci U S A. 2022. PMID: 36343267 Free PMC article.
-
Evolution of cancer stem cell lineage involving feedback regulation.PLoS One. 2021 May 20;16(5):e0251481. doi: 10.1371/journal.pone.0251481. eCollection 2021. PLoS One. 2021. PMID: 34014979 Free PMC article.
-
Kinesin Family Member 2A Serves as a Potential Biomarker Reflecting More Frequent Lymph Node Metastasis and Tumor Recurrence Risk in Basal-Like Breast Cancer Patients.Front Surg. 2022 Jun 16;9:889294. doi: 10.3389/fsurg.2022.889294. eCollection 2022. Front Surg. 2022. Retraction in: Front Surg. 2023 Oct 11;10:1307334. doi: 10.3389/fsurg.2023.1307334. PMID: 35784940 Free PMC article. Retracted.
-
Loss of Smek1 Induces Tauopathy and Triggers Neurodegeneration by Regulating Microtubule Stability.Adv Sci (Weinh). 2024 Oct;11(40):e2400584. doi: 10.1002/advs.202400584. Epub 2024 Aug 29. Adv Sci (Weinh). 2024. PMID: 39206808 Free PMC article.
-
Connecting neurodevelopment to neurodegeneration: a spotlight on the role of kinesin superfamily protein 2A (KIF2A).Neural Regen Res. 2024 Feb;19(2):375-379. doi: 10.4103/1673-5374.375298. Neural Regen Res. 2024. PMID: 37488893 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

