Blocking of Single α-Hemolysin Pore by Rhodamine Derivatives
- PMID: 28591605
- PMCID: PMC5474841
- DOI: 10.1016/j.bpj.2017.04.041
Blocking of Single α-Hemolysin Pore by Rhodamine Derivatives
Abstract
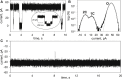
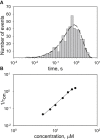
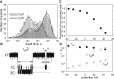
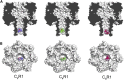
Measurements of ion conductance through α-hemolysin pore in a bilayer lipid membrane revealed blocking of the ion channel by a series of rhodamine 19 and rhodamine B esters. The longest dwell closed time of the blocking was observed with rhodamine 19 butyl ester (C4R1), whereas the octyl ester (C8R1) was of poor effect. Voltage asymmetry in the binding kinetics indicated that rhodamine derivatives bound to the stem part of the aqueous pore lumen. The binding frequency was proportional to a quadratic function of rhodamine concentrations, thereby showing that the dominant binding species were rhodamine dimers. Two levels of the pore conductance and two dwell closed times of the pore were found. The dwell closed times lengthened as the voltage increased, suggesting impermeability of the channel for the ligands. Molecular docking analysis revealed two distinct binding sites within the lumen of the stem of the α-hemolysin pore for the C4R1 dimer, but only one binding site for the C8R1 dimer. The blocking of the α-hemolysin nanopore by rhodamines could be utilized in DNA sequencing as additional optical sensing owing to bright fluorescence of rhodamines if used for DNA labeling.
Copyright © 2017 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Menestrina G. Ionic channels formed by Staphylococcus aureus α-toxin: voltage-dependent inhibition by divalent and trivalent cations. J. Membr. Biol. 1986;90:177–190. - PubMed
-
- Krasilnikov O.V., Sabirov R.Z. Ion transport through channels formed in lipid bilayers by Staphylococcus aureus α-toxin. Gen. Physiol. Biophys. 1989;8:213–222. - PubMed
-
- Song L., Hobaugh M.R., Gouaux J.E. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. - PubMed
-
- Bayley H., Cremer P.S. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

