Position-dependent effects of regioisomeric methylated adenine and guanine ribonucleosides on translation
- PMID: 28591780
- PMCID: PMC5587754
- DOI: 10.1093/nar/gkx515
Position-dependent effects of regioisomeric methylated adenine and guanine ribonucleosides on translation
Abstract
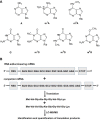
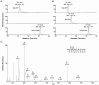
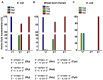
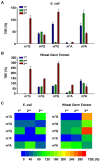
Reversible methylation of the N6 or N1 position of adenine in RNA has recently been shown to play significant roles in regulating the functions of RNA. RNA can also be alkylated upon exposure to endogenous and exogenous alkylating agents. Here we examined how regio-specific methylation at the hydrogen bonding edge of adenine and guanine in mRNA affects translation. When situated at the third codon position, the methylated nucleosides did not compromise the speed or accuracy of translation under most circumstances. When located at the first or second codon position, N1-methyladenosine (m1A) and m1G constituted robust blocks to both Escherichia coli and wheat germ extract translation systems, whereas N2-methylguanosine (m2G) moderately impeded translation. While m1A, m2G and N6-methyladenosine (m6A) did not perturb translational fidelity, O6-methylguanosine (m6G) at the first and second codon positions was strongly and moderately miscoding, respectively, and it was decoded as an adenosine in both systems. The effects of methylated ribonucleosides on translation could be attributed to the methylation-elicited alterations in base pairing properties of the nucleobases, and the mechanisms of ribosomal decoding contributed to the position-dependent effects. Together, our study afforded important new knowledge about the modulation of translation by methylation of purine nucleobases in mRNA.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
O6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity.RNA. 2015 Sep;21(9):1648-59. doi: 10.1261/rna.052464.115. Epub 2015 Jul 21. RNA. 2015. PMID: 26199454 Free PMC article.
-
N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.Nat Struct Mol Biol. 2016 Feb;23(2):110-5. doi: 10.1038/nsmb.3148. Epub 2016 Jan 11. Nat Struct Mol Biol. 2016. PMID: 26751643 Free PMC article.
-
Structural and Energetic Effects of O2'-Ribose Methylation of Protonated Purine Nucleosides.J Phys Chem B. 2018 Oct 4;122(39):9147-9160. doi: 10.1021/acs.jpcb.8b07687. Epub 2018 Sep 25. J Phys Chem B. 2018. PMID: 30203656
-
Mapping messenger RNA methylations at single base resolution.Curr Opin Chem Biol. 2021 Aug;63:28-37. doi: 10.1016/j.cbpa.2021.02.001. Epub 2021 Mar 5. Curr Opin Chem Biol. 2021. PMID: 33684855 Review.
-
Dependence of translation on 5'-terminal methylation of mRNA.Fed Proc. 1976 Sep;35(11):2214-7. Fed Proc. 1976. PMID: 782922 Review.
Cited by
-
Novel insight into the functions of N6‑methyladenosine modified lncRNAs in cancers (Review).Int J Oncol. 2022 Dec;61(6):152. doi: 10.3892/ijo.2022.5442. Epub 2022 Oct 20. Int J Oncol. 2022. PMID: 36263625 Free PMC article. Review.
-
Oxidation and alkylation stresses activate ribosome-quality control.Nat Commun. 2019 Dec 9;10(1):5611. doi: 10.1038/s41467-019-13579-3. Nat Commun. 2019. PMID: 31819057 Free PMC article.
-
How Messenger RNA and Nascent Chain Sequences Regulate Translation Elongation.Annu Rev Biochem. 2018 Jun 20;87:421-449. doi: 10.1146/annurev-biochem-060815-014818. Annu Rev Biochem. 2018. PMID: 29925264 Free PMC article. Review.
-
Genetic variants in m5C modification core genes are associated with the risk of Chinese pediatric acute lymphoblastic leukemia: A five-center case-control study.Front Oncol. 2023 Jan 9;12:1082525. doi: 10.3389/fonc.2022.1082525. eCollection 2022. Front Oncol. 2023. PMID: 36698387 Free PMC article.
-
Methylation of viral mRNA cap structures by PCIF1 attenuates the antiviral activity of interferon-β.Proc Natl Acad Sci U S A. 2021 Jul 20;118(29):e2025769118. doi: 10.1073/pnas.2025769118. Proc Natl Acad Sci U S A. 2021. PMID: 34266951 Free PMC article.
References
-
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. . Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. - PubMed
-
- Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C.. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 2016; 12:311–316. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

