Novel approaches in function-driven single-cell genomics
- PMID: 28591840
- PMCID: PMC5812545
- DOI: 10.1093/femsre/fux009
Novel approaches in function-driven single-cell genomics
Abstract
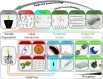
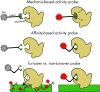
Deeper sequencing and improved bioinformatics in conjunction with single-cell and metagenomic approaches continue to illuminate undercharacterized environmental microbial communities. This has propelled the 'who is there, and what might they be doing' paradigm to the uncultivated and has already radically changed the topology of the tree of life and provided key insights into the microbial contribution to biogeochemistry. While characterization of 'who' based on marker genes can describe a large fraction of the community, answering 'what are they doing' remains the elusive pinnacle for microbiology. Function-driven single-cell genomics provides a solution by using a function-based screen to subsample complex microbial communities in a targeted manner for the isolation and genome sequencing of single cells. This enables single-cell sequencing to be focused on cells with specific phenotypic or metabolic characteristics of interest. Recovered genomes are conclusively implicated for both encoding and exhibiting the feature of interest, improving downstream annotation and revealing activity levels within that environment. This emerging approach has already improved our understanding of microbial community functioning and facilitated the experimental analysis of uncharacterized gene product space. Here we provide a comprehensive review of strategies that have been applied for function-driven single-cell genomics and the future directions we envision.
Keywords: function-driven sequencing; microbial dark matter; single-cell activity; single-cell genomics.
© FEMS 2017.
Figures

Similar articles
-
Microbial single-cell omics: the crux of the matter.Appl Microbiol Biotechnol. 2020 Oct;104(19):8209-8220. doi: 10.1007/s00253-020-10844-0. Epub 2020 Aug 26. Appl Microbiol Biotechnol. 2020. PMID: 32845367 Free PMC article. Review.
-
Co-cultivation of microbial sub-communities in microfluidic droplets facilitates high-resolution genomic dissection of microbial 'dark matter'.Integr Biol (Camb). 2020 Nov 18;12(11):263-274. doi: 10.1093/intbio/zyaa021. Integr Biol (Camb). 2020. PMID: 33089329 Free PMC article.
-
A single-cell genomics pipeline for environmental microbial eukaryotes.iScience. 2021 Mar 10;24(4):102290. doi: 10.1016/j.isci.2021.102290. eCollection 2021 Apr 23. iScience. 2021. PMID: 33870123 Free PMC article.
-
Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics.Nat Protoc. 2014 May;9(5):1038-48. doi: 10.1038/nprot.2014.067. Epub 2014 Apr 10. Nat Protoc. 2014. PMID: 24722403
-
[Progress in microbial single-cell genomics for environmental microbiology research - A review].Wei Sheng Wu Xue Bao. 2016 Nov 4;56(11):1691-8. Wei Sheng Wu Xue Bao. 2016. PMID: 29741831 Review. Chinese.
Cited by
-
Genome and pan-genome analysis to classify emerging bacteria.Biol Direct. 2019 Feb 26;14(1):5. doi: 10.1186/s13062-019-0234-0. Biol Direct. 2019. PMID: 30808378 Free PMC article. Review.
-
Next-generation physiology approaches to study microbiome function at single cell level.Nat Rev Microbiol. 2020 Apr;18(4):241-256. doi: 10.1038/s41579-020-0323-1. Epub 2020 Feb 13. Nat Rev Microbiol. 2020. PMID: 32055027 Free PMC article. Review.
-
Fluorometric Analysis of Carrier-Protein-Dependent Biosynthesis through a Conformationally Sensitive Solvatochromic Pantetheinamide Probe.ACS Chem Biol. 2024 Jul 19;19(7):1416-1425. doi: 10.1021/acschembio.4c00169. Epub 2024 Jun 23. ACS Chem Biol. 2024. PMID: 38909314 Free PMC article.
-
Microbial single-cell omics: the crux of the matter.Appl Microbiol Biotechnol. 2020 Oct;104(19):8209-8220. doi: 10.1007/s00253-020-10844-0. Epub 2020 Aug 26. Appl Microbiol Biotechnol. 2020. PMID: 32845367 Free PMC article. Review.
-
Targeted Cell Sorting Combined With Single Cell Genomics Captures Low Abundant Microbial Dark Matter With Higher Sensitivity Than Metagenomics.Front Microbiol. 2020 Jul 22;11:1377. doi: 10.3389/fmicb.2020.01377. eCollection 2020. Front Microbiol. 2020. PMID: 32793124 Free PMC article.
References
-
- Adam GC, Burbaum J, Kozarich JW et al. . Mapping enzyme active sites in complex proteomes. J Am Chem Soc 2004;126:1363–8. - PubMed
-
- Adam GC, Cravatt BF, Sorensen EJ. Profiling the specific reactivity of the proteome with non-directed activity-based probes. Chem Biol 2001;8:81–95. - PubMed
-
- Alderkamp AC, van Rijssel M, Bolhuis H. Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol Ecol 2007;59:108–17. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

