New insights into a classic aptamer: binding sites, cooperativity and more sensitive adenosine detection
- PMID: 28591844
- PMCID: PMC5737652
- DOI: 10.1093/nar/gkx517
New insights into a classic aptamer: binding sites, cooperativity and more sensitive adenosine detection
Abstract
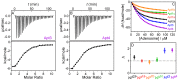
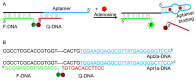
The DNA aptamer for adenosine (also for AMP and ATP) is a highly conserved sequence that has recurred in a few selections. It it a widely used model aptamer for biosensor development, and its nuclear magnetic resonance structure shows that each aptamer binds two AMP molecules. In this work, each binding site was individually removed by rational sequence design, while the remaining site still retained a similar binding affinity and specificity as confirmed by isothermal titration calorimetry. The thermodynamic parameters of binding are presented, and its biochemical implications are discussed. The number of binding sites can also be increased, and up to four sites are introduced in a single DNA sequence. Finally, the different sequences are made into fluorescent biosensors based on the structure-switching signaling aptamer design. The one-site aptamer has 3.8-fold higher sensitivity at lower adenosine concentration with a limit of detection of 9.1 μM adenosine, but weaker fluorescence signal at higher adenosine concentrations, consistent with a moderate cooperativity in the original aptamer. This work has offered insights into a classic aptamer for the relationship between the number of binding sites and sensitivity, and a shorter aptamer for improved biosensor design.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
A selective adenosine sensor derived from a triplex DNA aptamer.Anal Bioanal Chem. 2011 Jul;400(9):3035-40. doi: 10.1007/s00216-011-4996-1. Epub 2011 May 6. Anal Bioanal Chem. 2011. PMID: 21547431
-
Thermodynamic analysis of cooperative ligand binding by the ATP-binding DNA aptamer indicates a population-shift binding mechanism.Sci Rep. 2020 Nov 3;10(1):18944. doi: 10.1038/s41598-020-76002-8. Sci Rep. 2020. PMID: 33144644 Free PMC article.
-
Strategies for Creating Structure-Switching Aptamers.ACS Sens. 2018 Sep 28;3(9):1611-1615. doi: 10.1021/acssensors.8b00516. Epub 2018 Aug 29. ACS Sens. 2018. PMID: 30156834
-
Replacing antibodies with aptamers in lateral flow immunoassay.Biosens Bioelectron. 2015 Sep 15;71:230-242. doi: 10.1016/j.bios.2015.04.041. Epub 2015 Apr 14. Biosens Bioelectron. 2015. PMID: 25912679 Review.
-
Duplexed aptamers: history, design, theory, and application to biosensing.Chem Soc Rev. 2019 Mar 4;48(5):1390-1419. doi: 10.1039/c8cs00880a. Chem Soc Rev. 2019. PMID: 30707214 Review.
Cited by
-
Engineering base-excised aptamers for highly specific recognition of adenosine.Chem Sci. 2020 Feb 10;11(10):2735-2743. doi: 10.1039/d0sc00086h. Chem Sci. 2020. PMID: 34084332 Free PMC article.
-
Based on mutated aptamer-smartphone colorimetric detection of metronidazole in milk.Front Bioeng Biotechnol. 2024 Aug 2;12:1444846. doi: 10.3389/fbioe.2024.1444846. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39157440 Free PMC article.
-
Dissociation Constant (Kd) Measurement for Small-Molecule Binding Aptamers: Homogeneous Assay Methods and Critical Evaluations.Small Methods. 2025 Jun;9(6):e2401572. doi: 10.1002/smtd.202401572. Epub 2024 Nov 7. Small Methods. 2025. PMID: 39511863 Free PMC article. Review.
-
Cost-effective, user-friendly detection and preconcentration of thrombin on a sustainable paper-based electrochemical platform.Anal Bioanal Chem. 2025 Apr;417(9):1863-1872. doi: 10.1007/s00216-025-05764-9. Epub 2025 Feb 1. Anal Bioanal Chem. 2025. PMID: 39891658 Free PMC article.
-
Genetically Encoded Fluorogenic DNA Aptamers for Imaging Metabolite in Living Cells.J Am Chem Soc. 2025 Jan 15;147(2):1529-1541. doi: 10.1021/jacs.4c09855. Epub 2024 Dec 31. J Am Chem Soc. 2025. PMID: 39739942 Free PMC article.
References
-
- Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. - PubMed
-
- Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. - PubMed
-
- Winkler W., Nahvi A., Breaker R.R.. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002; 419:952–956. - PubMed
-
- Winkler W.C., Breaker R.R.. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005; 59:487–517. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

