Genomes of the Mouse Collaborative Cross
- PMID: 28592495
- PMCID: PMC5499171
- DOI: 10.1534/genetics.116.198838
Genomes of the Mouse Collaborative Cross
Abstract
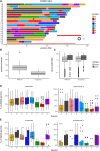
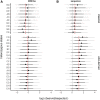
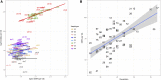
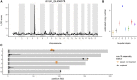
The Collaborative Cross (CC) is a multiparent panel of recombinant inbred (RI) mouse strains derived from eight founder laboratory strains. RI panels are popular because of their long-term genetic stability, which enhances reproducibility and integration of data collected across time and conditions. Characterization of their genomes can be a community effort, reducing the burden on individual users. Here we present the genomes of the CC strains using two complementary approaches as a resource to improve power and interpretation of genetic experiments. Our study also provides a cautionary tale regarding the limitations imposed by such basic biological processes as mutation and selection. A distinct advantage of inbred panels is that genotyping only needs to be performed on the panel, not on each individual mouse. The initial CC genome data were haplotype reconstructions based on dense genotyping of the most recent common ancestors (MRCAs) of each strain followed by imputation from the genome sequence of the corresponding founder inbred strain. The MRCA resource captured segregating regions in strains that were not fully inbred, but it had limited resolution in the transition regions between founder haplotypes, and there was uncertainty about founder assignment in regions of limited diversity. Here we report the whole genome sequence of 69 CC strains generated by paired-end short reads at 30× coverage of a single male per strain. Sequencing leads to a substantial improvement in the fine structure and completeness of the genomes of the CC. Both MRCAs and sequenced samples show a significant reduction in the genome-wide haplotype frequencies from two wild-derived strains, CAST/EiJ and PWK/PhJ. In addition, analysis of the evolution of the patterns of heterozygosity indicates that selection against three wild-derived founder strains played a significant role in shaping the genomes of the CC. The sequencing resource provides the first description of tens of thousands of new genetic variants introduced by mutation and drift in the CC genomes. We estimate that new SNP mutations are accumulating in each CC strain at a rate of 2.4 ± 0.4 per gigabase per generation. The fixation of new mutations by genetic drift has introduced thousands of new variants into the CC strains. The majority of these mutations are novel compared to currently sequenced laboratory stocks and wild mice, and some are predicted to alter gene function. Approximately one-third of the CC inbred strains have acquired large deletions (>10 kb) many of which overlap known coding genes and functional elements. The sequence of these mice is a critical resource to CC users, increases threefold the number of mouse inbred strain genomes available publicly, and provides insight into the effect of mutation and drift on common resources.
Keywords: MPP; drift; genetic variants; multiparental populations; selection; whole genome sequence.
Copyright © 2017 Srivastava et al.
Figures



Similar articles
-
Male Infertility Is Responsible for Nearly Half of the Extinction Observed in the Mouse Collaborative Cross.Genetics. 2017 Jun;206(2):557-572. doi: 10.1534/genetics.116.199596. Genetics. 2017. PMID: 28592496 Free PMC article.
-
The genome architecture of the Collaborative Cross mouse genetic reference population.Genetics. 2012 Feb;190(2):389-401. doi: 10.1534/genetics.111.132639. Genetics. 2012. PMID: 22345608 Free PMC article.
-
Joint Analysis of Strain and Parent-of-Origin Effects for Recombinant Inbred Intercrosses Generated from Multiparent Populations with the Collaborative Cross as an Example.G3 (Bethesda). 2018 Feb 2;8(2):599-605. doi: 10.1534/g3.117.300483. G3 (Bethesda). 2018. PMID: 29255115 Free PMC article.
-
The Collaborative Cross mouse model for dissecting genetic susceptibility to infectious diseases.Mamm Genome. 2018 Aug;29(7-8):471-487. doi: 10.1007/s00335-018-9768-1. Epub 2018 Aug 24. Mamm Genome. 2018. PMID: 30143822 Review.
-
A novel strategy for genetic dissection of complex traits: the population of specific chromosome substitution strains from laboratory and wild mice.Mamm Genome. 2010 Aug;21(7-8):370-6. doi: 10.1007/s00335-010-9270-x. Epub 2010 Jul 11. Mamm Genome. 2010. PMID: 20623355 Review.
Cited by
-
Companion Animals as Models for Inhibition of STAT3 and STAT5.Cancers (Basel). 2019 Dec 17;11(12):2035. doi: 10.3390/cancers11122035. Cancers (Basel). 2019. PMID: 31861073 Free PMC article. Review.
-
Host-pathogen genetic interactions underlie tuberculosis susceptibility in genetically diverse mice.Elife. 2022 Feb 3;11:e74419. doi: 10.7554/eLife.74419. Elife. 2022. PMID: 35112666 Free PMC article.
-
ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs.Nature. 2022 Jun;606(7913):358-367. doi: 10.1038/s41586-022-04769-z. Epub 2022 Apr 27. Nature. 2022. PMID: 35477154 Free PMC article.
-
A diversity outbred F1 mouse model identifies host-intrinsic genetic regulators of response to immune checkpoint inhibitors.Oncoimmunology. 2022 Apr 20;11(1):2064958. doi: 10.1080/2162402X.2022.2064958. eCollection 2022. Oncoimmunology. 2022. PMID: 35481286 Free PMC article.
-
Diverse CD8 T Cell Responses to Viral Infection Revealed by the Collaborative Cross.Cell Rep. 2020 Apr 14;31(2):107508. doi: 10.1016/j.celrep.2020.03.072. Cell Rep. 2020. PMID: 32294433 Free PMC article.
References
-
- Bouquet M., Selva J., Auroux M., 1993. Cryopreservation of mouse oocytes: Mutagenic effects in the embryo? Biol. Reprod. 49: 764–769. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

