Epistatic Networks Jointly Influence Phenotypes Related to Metabolic Disease and Gene Expression in Diversity Outbred Mice
- PMID: 28592500
- PMCID: PMC5499176
- DOI: 10.1534/genetics.116.198051
Epistatic Networks Jointly Influence Phenotypes Related to Metabolic Disease and Gene Expression in Diversity Outbred Mice
Abstract
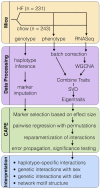
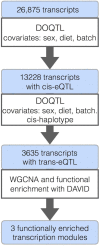
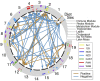
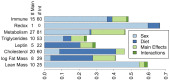
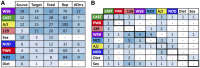
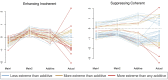
Genetic studies of multidimensional phenotypes can potentially link genetic variation, gene expression, and physiological data to create multi-scale models of complex traits. The challenge of reducing these data to specific hypotheses has become increasingly acute with the advent of genome-scale data resources. Multi-parent populations derived from model organisms provide a resource for developing methods to understand this complexity. In this study, we simultaneously modeled body composition, serum biomarkers, and liver transcript abundances from 474 Diversity Outbred mice. This population contained both sexes and two dietary cohorts. Transcript data were reduced to functional gene modules with weighted gene coexpression network analysis (WGCNA), which were used as summary phenotypes representing enriched biological processes. These module phenotypes were jointly analyzed with body composition and serum biomarkers in a combined analysis of pleiotropy and epistasis (CAPE), which inferred networks of epistatic interactions between quantitative trait loci that affect one or more traits. This network frequently mapped interactions between alleles of different ancestries, providing evidence of both genetic synergy and redundancy between haplotypes. Furthermore, a number of loci interacted with sex and diet to yield sex-specific genetic effects and alleles that potentially protect individuals from the effects of a high-fat diet. Although the epistatic interactions explained small amounts of trait variance, the combination of directional interactions, allelic specificity, and high genomic resolution provided context to generate hypotheses for the roles of specific genes in complex traits. Our approach moves beyond the cataloging of single loci to infer genetic networks that map genetic etiology by simultaneously modeling all phenotypes.
Keywords: MPP; epistasis; multiparental populations; outbred mouse population; pleiotropy; systems genetics.
Copyright © 2017 by the Genetics Society of America.
Figures









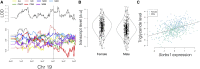
Similar articles
-
The Combined Analysis of Pleiotropy and Epistasis (CAPE).Methods Mol Biol. 2021;2212:55-67. doi: 10.1007/978-1-0716-0947-7_5. Methods Mol Biol. 2021. PMID: 33733350
-
Dissection of complex gene expression using the combined analysis of pleiotropy and epistasis.Pac Symp Biocomput. 2014:200-11. Pac Symp Biocomput. 2014. PMID: 24297548 Free PMC article.
-
CAPE: an R package for combined analysis of pleiotropy and epistasis.PLoS Comput Biol. 2013 Oct;9(10):e1003270. doi: 10.1371/journal.pcbi.1003270. Epub 2013 Oct 24. PLoS Comput Biol. 2013. PMID: 24204223 Free PMC article.
-
Epistasis and pleiotropy-induced variation for plant breeding.Plant Biotechnol J. 2024 Oct;22(10):2788-2807. doi: 10.1111/pbi.14405. Epub 2024 Jun 14. Plant Biotechnol J. 2024. PMID: 38875130 Free PMC article. Review.
-
The detection and characterization of pleiotropy: discovery, progress, and promise.Brief Bioinform. 2016 Jan;17(1):13-22. doi: 10.1093/bib/bbv050. Epub 2015 Jul 28. Brief Bioinform. 2016. PMID: 26223525 Review.
Cited by
-
Prediction performance of linear models and gradient boosting machine on complex phenotypes in outbred mice.G3 (Bethesda). 2022 Apr 4;12(4):jkac039. doi: 10.1093/g3journal/jkac039. G3 (Bethesda). 2022. PMID: 35166767 Free PMC article.
-
The potential of integrating human and mouse discovery platforms to advance our understanding of cardiometabolic diseases.Elife. 2023 Mar 31;12:e86139. doi: 10.7554/eLife.86139. Elife. 2023. PMID: 37000167 Free PMC article.
-
Epistasis: Searching for Interacting Genetic Variants Using Crosses.G3 (Bethesda). 2017 Jun 7;7(6):1619-1622. doi: 10.1534/g3.117.042770. G3 (Bethesda). 2017. PMID: 28592644 Free PMC article. No abstract available.
-
Natural genetic variation in wild-derived mice controls host survival and transcriptional responses during endotoxic shock.Immunohorizons. 2025 Mar 26;9(5):vlaf007. doi: 10.1093/immhor/vlaf007. Immunohorizons. 2025. PMID: 40139977 Free PMC article.
-
Hepatic transcriptional profile reveals the role of diet and genetic backgrounds on metabolic traits in female progenitor strains of the Collaborative Cross.Physiol Genomics. 2021 May 1;53(5):173-192. doi: 10.1152/physiolgenomics.00140.2020. Epub 2021 Apr 5. Physiol Genomics. 2021. PMID: 33818129 Free PMC article.
References
-
- Agrawal H., 2002. Extreme self-organization in networks constructed from gene expression data. Phys. Rev. Lett. 89: 268702. - PubMed
-
- Albert F. W., Kruglyak L., 2015. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. - PubMed
-
- Barabási A.-L., Oltvai Z. N., 2004. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5: 101–113. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

