Whole-Genome Resequencing Identifies the Molecular Genetic Cause for the Absence of a Gy5 Glycinin Protein in Soybean PI 603408
- PMID: 28592556
- PMCID: PMC5499141
- DOI: 10.1534/g3.117.039347
Whole-Genome Resequencing Identifies the Molecular Genetic Cause for the Absence of a Gy5 Glycinin Protein in Soybean PI 603408
Abstract
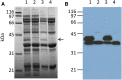
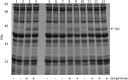
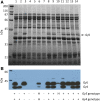
During ongoing proteomic analysis of the soybean (Glycine max (L.) Merr) germplasm collection, PI 603408 was identified as a landrace whose seeds lack accumulation of one of the major seed storage glycinin protein subunits. Whole genomic resequencing was used to identify a two-base deletion affecting glycinin 5 The newly discovered deletion was confirmed to be causative through immunological, genetic, and proteomic analysis, and no significant differences in total seed protein content were found to be due to the glycinin 5 loss-of-function mutation per se In addition to focused studies on this one specific glycinin subunit-encoding gene, a total of 1,858,185 nucleotide variants were identified, of which 39,344 were predicted to affect protein coding regions. In order to semiautomate analysis of a large number of soybean gene variants, a new SIFT 4G (Sorting Intolerant From Tolerated 4 Genomes) database was designed to predict the impact of nonsynonymous single nucleotide soybean gene variants, potentially enabling more rapid analysis of soybean resequencing data in the future.
Keywords: Glycine max; amino acid changes (AAC); genomic resequencing; glycinin; seed proteomics; single nucleotide variants (SNV); soybean.
Copyright © 2017 Gillman et al.
Figures

Similar articles
-
Linkage and association study discovered loci and candidate genes for glycinin and β-conglycinin in soybean (Glycine max L. Merr.).Theor Appl Genet. 2021 Apr;134(4):1201-1215. doi: 10.1007/s00122-021-03766-6. Epub 2021 Jan 19. Theor Appl Genet. 2021. PMID: 33464377
-
Quantitative Trait Loci (QTL) Mapping for Glycinin and β-Conglycinin Contents in Soybean (Glycine max L. Merr.).J Agric Food Chem. 2016 May 4;64(17):3473-83. doi: 10.1021/acs.jafc.6b00167. Epub 2016 Apr 25. J Agric Food Chem. 2016. PMID: 27070305
-
Proteomic and genetic analysis of glycinin subunits of sixteen soybean genotypes.Plant Physiol Biochem. 2007 Jun-Jul;45(6-7):436-44. doi: 10.1016/j.plaphy.2007.03.031. Epub 2007 Apr 6. Plant Physiol Biochem. 2007. PMID: 17524657
-
Structural and functional analysis of various globulin proteins from soy seed.Crit Rev Food Sci Nutr. 2015;55(11):1491-502. doi: 10.1080/10408398.2012.700340. Crit Rev Food Sci Nutr. 2015. PMID: 24915310 Review.
-
Update soybean Zhonghuang 13 genome to a golden reference.Sci China Life Sci. 2019 Sep;62(9):1257-1260. doi: 10.1007/s11427-019-9822-2. Epub 2019 Aug 21. Sci China Life Sci. 2019. PMID: 31444683 Review. No abstract available.
Cited by
-
Molecular characterization of genomic regions for resistance to Pythium ultimum var. ultimum in the soybean cultivar Magellan.Theor Appl Genet. 2019 Feb;132(2):405-417. doi: 10.1007/s00122-018-3228-x. Epub 2018 Nov 15. Theor Appl Genet. 2019. PMID: 30443655
References
-
- Beilinson V., Chen Z., Shoemaker R., Fischer R., Goldberg R., et al. , 2002. Genomic organization of glycinin genes in soybean. Theor. Appl. Genet. 104(6): 1132–1140. - PubMed
-
- Coates J. B., Medeiros J. S., Thanh V. H., Nielsen N. C., 1985. Characterization of the subunits of β-conglycinin. Arch. Biochem. Biophys. 243(1): 184–194. - PubMed
-
- Fang C., Li C., Li W., Wang Z., Zhou Z., et al. , 2014. Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. Plant J. 77(5): 700–712. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
