Rear-polarized Wnt5a-receptor-actin-myosin-polarity (WRAMP) structures promote the speed and persistence of directional cell migration
- PMID: 28592632
- PMCID: PMC5541843
- DOI: 10.1091/mbc.E16-12-0875
Rear-polarized Wnt5a-receptor-actin-myosin-polarity (WRAMP) structures promote the speed and persistence of directional cell migration
Abstract
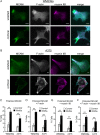
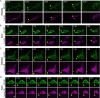
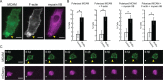
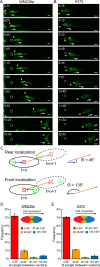
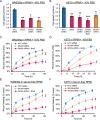
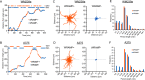
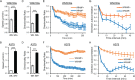
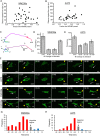
In contrast to events at the cell leading edge, rear-polarized mechanisms that control directional cell migration are poorly defined. Previous work described a new intracellular complex, the Wnt5a-receptor-actomyosin polarity (WRAMP) structure, which coordinates the polarized localization of MCAM, actin, and myosin IIB in a Wnt5a-induced manner. However, the polarity and function for the WRAMP structure during cell movement were not determined. Here we characterize WRAMP structures during extended cell migration using live-cell imaging. The results demonstrate that cells undergoing prolonged migration show WRAMP structures stably polarized at the rear, where they are strongly associated with enhanced speed and persistence of directional movement. Strikingly, WRAMP structures form transiently, with cells displaying directional persistence during periods when they are present and cells changing directions randomly when they are absent. Cells appear to pause locomotion when WRAMP structures disassemble and then migrate in new directions after reassembly at a different location, which forms the new rear. We conclude that WRAMP structures represent a rear-directed cellular mechanism to control directional migration and that their ability to form dynamically within cells may control changes in direction during extended migration.
© 2017 Connacher et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
References
-
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. - PubMed
-
- Cramer LP. Forming the cell rear first: breaking cell symmetry to trigger directed cell migration. Nat Cell Biol. 2010;12:628–632. - PubMed
-
- Cramer LP. Mechanism of cell rear retraction in migrating cells. Curr Opin Cell Biol. 2013;25:591–599. - PubMed
-
- Gao B. Wnt regulation of planar cell polarity (PCP) Curr Top Dev Biol. 2012;101:263–295. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

