First comprehensive proteome analysis of lysine crotonylation in seedling leaves of Nicotiana tabacum
- PMID: 28592803
- PMCID: PMC5462846
- DOI: 10.1038/s41598-017-03369-6
First comprehensive proteome analysis of lysine crotonylation in seedling leaves of Nicotiana tabacum
Abstract
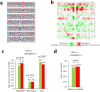
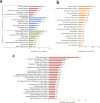
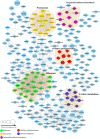
Histone crotonylation is a new lysine acylation type of post-translational modification (PTM) enriched at active gene promoters and potential enhancers in yeast and mammalian cells. However, lysine crotonylation in nonhistone proteins and plant cells has not yet been studied. In the present study, we performed a global crotonylation proteome analysis of Nicotiana tabacum (tobacco) using high-resolution LC-MS/MS coupled with highly sensitive immune-affinity purification. A total of 2044 lysine modification sites distributed on 637 proteins were identified, representing the most abundant lysine acylation proteome reported in the plant kingdom. Similar to lysine acetylation and succinylation in plants, lysine crotonylation was related to multiple metabolism pathways, such as carbon metabolism, the citrate cycle, glycolysis, and the biosynthesis of amino acids. Importantly, 72 proteins participated in multiple processes of photosynthesis, and most of the enzymes involved in chlorophyll synthesis were modified through crotonylation. Numerous crotonylated proteins were implicated in the biosynthesis, folding, and degradation of proteins through the ubiquitin-proteasome system. Several crotonylated proteins related to chromatin organization are also discussed here. These data represent the first report of a global crotonylation proteome and provide a promising starting point for further functional research of crotonylation in nonhistone proteins.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
A qualitative proteome-wide lysine crotonylation profiling of papaya (Carica papaya L.).Sci Rep. 2018 May 29;8(1):8230. doi: 10.1038/s41598-018-26676-y. Sci Rep. 2018. PMID: 29844531 Free PMC article.
-
Ammonium triggered the response mechanism of lysine crotonylome in tea plants.BMC Genomics. 2019 May 6;20(1):340. doi: 10.1186/s12864-019-5716-z. BMC Genomics. 2019. PMID: 31060518 Free PMC article.
-
Comprehensive analysis of lysine crotonylation in proteome of maintenance hemodialysis patients.Medicine (Baltimore). 2018 Sep;97(37):e12035. doi: 10.1097/MD.0000000000012035. Medicine (Baltimore). 2018. PMID: 30212933 Free PMC article.
-
Functions and mechanisms of lysine crotonylation.J Cell Mol Med. 2019 Nov;23(11):7163-7169. doi: 10.1111/jcmm.14650. Epub 2019 Sep 1. J Cell Mol Med. 2019. PMID: 31475443 Free PMC article. Review.
-
Lysine crotonylation: A challenging new player in the epigenetic regulation of plants.J Proteomics. 2022 Mar 20;255:104488. doi: 10.1016/j.jprot.2022.104488. Epub 2022 Jan 20. J Proteomics. 2022. PMID: 35065287 Review.
Cited by
-
Qualitative lysine crotonylome analysis in the ovarian tissue of Harmonia axyridis (Pallas).PLoS One. 2021 Oct 18;16(10):e0258371. doi: 10.1371/journal.pone.0258371. eCollection 2021. PLoS One. 2021. PMID: 34662345 Free PMC article.
-
Large-Scale Identification of Lysine Crotonylation Reveals Its Potential Role in Oral Squamous Cell Carcinoma.Cancer Manag Res. 2023 Oct 17;15:1165-1179. doi: 10.2147/CMAR.S424422. eCollection 2023. Cancer Manag Res. 2023. PMID: 37868687 Free PMC article.
-
Protein lysine crotonylation in cellular processions and disease associations.Genes Dis. 2023 Aug 2;11(5):101060. doi: 10.1016/j.gendis.2023.06.029. eCollection 2024 Sep. Genes Dis. 2023. PMID: 38957707 Free PMC article. Review.
-
Lysine crotonylation of DgTIL1 at K72 modulates cold tolerance by enhancing DgnsLTP stability in chrysanthemum.Plant Biotechnol J. 2021 Jun;19(6):1125-1140. doi: 10.1111/pbi.13533. Epub 2021 Jan 21. Plant Biotechnol J. 2021. PMID: 33368971 Free PMC article.
-
Cataloging Posttranslational Modifications in Plant Histones.Adv Exp Med Biol. 2021;1346:131-154. doi: 10.1007/978-3-030-80352-0_8. Adv Exp Med Biol. 2021. PMID: 35113400
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

