Activity-independent targeting of mTOR to lysosomes in primary osteoclasts
- PMID: 28592812
- PMCID: PMC5462732
- DOI: 10.1038/s41598-017-03494-2
Activity-independent targeting of mTOR to lysosomes in primary osteoclasts
Abstract
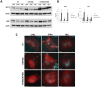
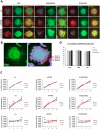
Mammalian target of rapamycin (mTOR) is activated by numerous stimuli, including amino acids and growth factors. This kinase is part of the mTOR complex 1 (mTORC1) which regulates cell proliferation, differentiation, and autophagy. Active mTORC1 is located on lysosomes and has been reported to disassociate from the lysosomal surface in the absence of amino acids. Furthermore, mTORC1 activity has been linked to the vacuolar H+-ATPases (V-ATPases), the proton pumps responsible for lysosomal acidification; however, the exact role of the V-ATPases in mTORC1 signaling is not known. To elucidate the mechanisms involved in mTORC1 regulation by the V-ATPases, we used primary osteoclasts derived from mice carrying a point (R740S) mutation in the a3 subunit of the V-ATPase. In these cells, the mutant protein is expressed but the pump is not functional, resulting in higher lysosomal pH. By analyzing mTOR activation, mTOR/lysosome co-localization, and lysosomal positioning using confocal microscopy, fractionation, and ultrapure lysosomal purification methods, we demonstrate that in primary osteoclasts, mTOR is localized on the lysosomal surface even when mTOR activity is inhibited. Our findings reveal that mTOR targeting to the lysosome in osteoclasts is activity-independent, and that its disassociation from the lysosome during starvation is not universal.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

