Quantitative Proteomics Shows Extensive Remodeling Induced by Nitrogen Limitation in Prochlorococcusmarinus SS120
- PMID: 28593196
- PMCID: PMC5451487
- DOI: 10.1128/mSystems.00008-17
Quantitative Proteomics Shows Extensive Remodeling Induced by Nitrogen Limitation in Prochlorococcusmarinus SS120
Abstract
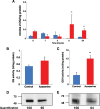
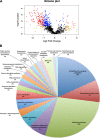
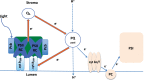
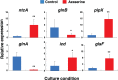
Prochlorococcus requires the capability to accommodate to environmental changes in order to proliferate in oligotrophic oceans, in particular regarding nitrogen availability. A precise knowledge of the composition and changes in the proteome can yield fundamental insights into such a response. Here we report a detailed proteome analysis of the important model cyanobacterium Prochlorococcus marinus SS120 after treatment with azaserine, an inhibitor of ferredoxin-dependent glutamate synthase (GOGAT), to simulate extreme nitrogen starvation. In total, 1,072 proteins, corresponding to 57% of the theoretical proteome, were identified-the maximum proteome coverage obtained for any Prochlorococcus strain thus far. Spectral intensity, calibrated quantification by the Hi3 method, was obtained for 1,007 proteins. Statistically significant changes (P value of <0.05) were observed for 408 proteins, with the majority of proteins (92.4%) downregulated after 8 h of treatment. There was a strong decrease in ribosomal proteins upon azaserine addition, while many transporters were increased. The regulatory proteins PII and PipX were decreased, and the global nitrogen regulator NtcA was upregulated. Furthermore, our data for Prochlorococcus indicate that NtcA also participates in the regulation of photosynthesis. Prochlorococcus responds to the lack of nitrogen by slowing down translation, while inducing photosynthetic cyclic electron flow and biosynthesis of proteins involved in nitrogen uptake and assimilation. IMPORTANCEProchlorococcus is the most abundant photosynthetic organism on Earth, contributing significantly to global primary production and playing a prominent role in biogeochemical cycles. Here we study the effects of extreme nitrogen limitation, a feature of the oligotrophic oceans inhabited by this organism. Quantitative proteomics allowed an accurate quantification of the Prochlorococcus proteome, finding three main responses to nitrogen limitation: upregulation of nitrogen assimilation-related proteins, including transporters; downregulation of ribosome proteins; and induction of the photosystem II cyclic electron flow. This suggests that nitrogen limitation affects a range of metabolic processes far wider than initially believed, with the ultimate goal of saving nitrogen and maximizing the nitrogen uptake and assimilation capabilities of the cell.
Keywords: marine cyanobacteria; nitrogen limitation; nitrogen metabolism; prochlorococcus; quantitative proteomics.
Figures

References
-
- Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, West-Johnsrud L, Zettler ER. 1992. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and chlorophyll b. Arch Microbiol 157:297–300. doi: 10.1007/BF00245165. - DOI
-
- Chisholm SW, Olson RJ, Zettler ER, Goericke R, Waterbury JB, Welschmeyer NA. 1988. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334:340–343. doi: 10.1038/334340a0. - DOI
-
- Liu H, Nolla H, Campbell L. 1997. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat Microb Ecol 12:39–47. doi: 10.3354/ame012039. - DOI
-
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, Chen F, Lapidus A, Ferriera S, Johnson J, Steglich C, Church GM, Richardson P, Chisholm SW. 2007. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3:e231. doi: 10.1371/journal.pgen.0030231. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
