Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis
- PMID: 28593988
- PMCID: PMC5472179
- DOI: 10.1038/ncomms15576
Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis
Abstract
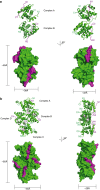
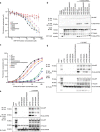
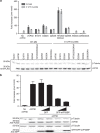
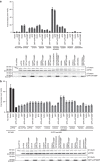
Ebola virus (EBOV) is an enveloped negative-sense RNA virus that causes sporadic outbreaks with high case fatality rates. Ebola viral protein 30 (eVP30) plays a critical role in EBOV transcription initiation at the nucleoprotein (eNP) gene, with additional roles in the replication cycle such as viral assembly. However, the mechanistic basis for how eVP30 functions during the virus replication cycle is currently unclear. Here we define a key interaction between eVP30 and a peptide derived from eNP that is important to facilitate interactions leading to the recognition of the RNA template. We present crystal structures of the eVP30 C-terminus in complex with this eNP peptide. Functional analyses of the eVP30-eNP interface identify residues that are critical for viral RNA synthesis. Altogether, these results support a model where the eVP30-eNP interaction plays a critical role in transcription initiation and provides a novel target for the development of antiviral therapy.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Virus and host interactions critical for filoviral RNA synthesis as therapeutic targets.Antiviral Res. 2019 Feb;162:90-100. doi: 10.1016/j.antiviral.2018.12.006. Epub 2018 Dec 11. Antiviral Res. 2019. PMID: 30550800 Free PMC article. Review.
-
Serine-Arginine Protein Kinase 1 Regulates Ebola Virus Transcription.mBio. 2020 Feb 25;11(1):e02565-19. doi: 10.1128/mBio.02565-19. mBio. 2020. PMID: 32098814 Free PMC article.
-
RNA Binding of Ebola Virus VP30 Is Essential for Activating Viral Transcription.J Virol. 2016 Jul 27;90(16):7481-7496. doi: 10.1128/JVI.00271-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27279615 Free PMC article.
-
Dynamic Phosphorylation of VP30 Is Essential for Ebola Virus Life Cycle.J Virol. 2016 Apr 29;90(10):4914-4925. doi: 10.1128/JVI.03257-15. Print 2016 May 15. J Virol. 2016. PMID: 26937028 Free PMC article.
-
Structural insights into the rhabdovirus transcription/replication complex.Virus Res. 2011 Dec;162(1-2):126-37. doi: 10.1016/j.virusres.2011.09.025. Epub 2011 Sep 22. Virus Res. 2011. PMID: 21963663 Review.
Cited by
-
Virus and host interactions critical for filoviral RNA synthesis as therapeutic targets.Antiviral Res. 2019 Feb;162:90-100. doi: 10.1016/j.antiviral.2018.12.006. Epub 2018 Dec 11. Antiviral Res. 2019. PMID: 30550800 Free PMC article. Review.
-
Regulation of VP30-Dependent Transcription by RNA Sequence and Structure in the Genomic Ebola Virus Promoter.J Virol. 2021 Mar 1;95(5):e02215-20. doi: 10.1128/JVI.02215-20. Epub 2020 Dec 2. J Virol. 2021. PMID: 33268520 Free PMC article.
-
Serine-arginine protein kinases and their targets in viral infection and their inhibition.Cell Mol Life Sci. 2023 May 17;80(6):153. doi: 10.1007/s00018-023-04808-6. Cell Mol Life Sci. 2023. PMID: 37198350 Free PMC article. Review.
-
Serine-Arginine Protein Kinase 1 Regulates Ebola Virus Transcription.mBio. 2020 Feb 25;11(1):e02565-19. doi: 10.1128/mBio.02565-19. mBio. 2020. PMID: 32098814 Free PMC article.
-
Small molecule drug discovery for Ebola virus disease.RSC Med Chem. 2025 Aug 6. doi: 10.1039/d5md00533g. Online ahead of print. RSC Med Chem. 2025. PMID: 40852580 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

