Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge
- PMID: 28593989
- PMCID: PMC5472724
- DOI: 10.1038/ncomms15711
Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge
Abstract
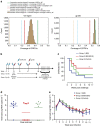
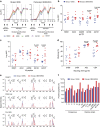
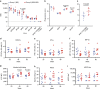
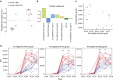
The RV144 Thai trial HIV-1 vaccine of recombinant poxvirus (ALVAC) and recombinant HIV-1 gp120 subtype B/subtype E (B/E) proteins demonstrated 31% vaccine efficacy. Here we design an ALVAC/Pentavalent B/E/E/E/E vaccine to increase the diversity of gp120 motifs in the immunogen to elicit a broader antibody response and enhance protection. We find that immunization of rhesus macaques with the pentavalent vaccine results in protection of 55% of pentavalent-vaccine-immunized macaques from simian-human immunodeficiency virus (SHIV) challenge. Systems serology of the antibody responses identifies plasma antibody binding to HIV-infected cells, peak ADCC antibody titres, NK cell-mediated ADCC and antibody-mediated activation of MIP-1β in NK cells as the four immunological parameters that best predict decreased infection risk that are improved by the pentavalent vaccine. Thus inclusion of additional gp120 immunogens to a pox-prime/protein boost regimen can augment antibody responses and enhance protection from a SHIV challenge in rhesus macaques.
Conflict of interest statement
S.P. and J.T. are employees of Sanofi Pasteur. B.T.K. and B.F.H. have patent applications submitted on vaccine candidates used in this study. The remaining authors declare no competing financial interests.
Figures

References
-
- Rerks-Ngarm S. et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361, 2209–2220 (2009). - PubMed
-
- Flynn N. M. et al. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191, 654–665 (2005). - PubMed
-
- Pitisuttithum P. et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 194, 1661–1671 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

