Interactive Roles of DNA Helicases and Translocases with the Single-Stranded DNA Binding Protein RPA in Nucleic Acid Metabolism
- PMID: 28594346
- PMCID: PMC5486056
- DOI: 10.3390/ijms18061233
Interactive Roles of DNA Helicases and Translocases with the Single-Stranded DNA Binding Protein RPA in Nucleic Acid Metabolism
Abstract
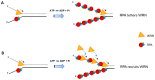
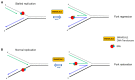
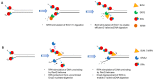
Helicases and translocases use the energy of nucleoside triphosphate binding and hydrolysis to unwind/resolve structured nucleic acids or move along a single-stranded or double-stranded polynucleotide chain, respectively. These molecular motors facilitate a variety of transactions including replication, DNA repair, recombination, and transcription. A key partner of eukaryotic DNA helicases/translocases is the single-stranded DNA binding protein Replication Protein A (RPA). Biochemical, genetic, and cell biological assays have demonstrated that RPA interacts with these human molecular motors physically and functionally, and their association is enriched in cells undergoing replication stress. The roles of DNA helicases/translocases are orchestrated with RPA in pathways of nucleic acid metabolism. RPA stimulates helicase-catalyzed DNA unwinding, enlists translocases to sites of action, and modulates their activities in DNA repair, fork remodeling, checkpoint activation, and telomere maintenance. The dynamic interplay between DNA helicases/translocases and RPA is just beginning to be understood at the molecular and cellular levels, and there is still much to be learned, which may inform potential therapeutic strategies.
Keywords: DNA repair; RPA; Replication Protein A; checkpoint; helicase; replication; telomere; translocase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

