Comparing Proteolytic Fingerprints of Antigen-Presenting Cells during Allergen Processing
- PMID: 28594355
- PMCID: PMC5486048
- DOI: 10.3390/ijms18061225
Comparing Proteolytic Fingerprints of Antigen-Presenting Cells during Allergen Processing
Abstract
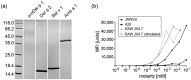
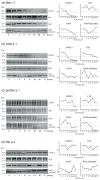
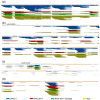
Endolysosomal processing has a critical influence on immunogenicity as well as immune polarization of protein antigens. In industrialized countries, allergies affect around 25% of the population. For the rational design of protein-based allergy therapeutics for immunotherapy, a good knowledge of T cell-reactive regions on allergens is required. Thus, we sought to analyze endolysosomal degradation patterns of inhalant allergens. Four major allergens from ragweed, birch, as well as house dust mites were produced as recombinant proteins. Endolysosomal proteases were purified by differential centrifugation from dendritic cells, macrophages, and B cells, and combined with allergens for proteolytic processing. Thereafter, endolysosomal proteolysis was monitored by protein gel electrophoresis and mass spectrometry. We found that the overall proteolytic activity of specific endolysosomal fractions differed substantially, whereas the degradation patterns of the four model allergens obtained with the different proteases were extremely similar. Moreover, previously identified T cell epitopes were assigned to endolysosomal peptides and indeed showed a good overlap with known T cell epitopes for all four candidate allergens. Thus, we propose that the degradome assay can be used as a predictor to determine antigenic peptides as potential T cell epitopes, which will help in the rational design of protein-based allergy vaccine candidates.
Keywords: Amb a 1; Bet v 1; Der p 1; Der p 2; allergen proteolysis; degradome assay; proteases from B cells; proteases from dendritic cells; proteases from macrophages.
Conflict of interest statement
Michael Wallner received funding from the Austrian Science Funds FWF, the University of Salzburg, OeAD-GmbH, and Sparkling Science. Teresa Twaroch, Frank Stolz, and Angela Neubauer are or were employees of Biomay AG. All other authors declare no possible conflict of interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

