Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways
- PMID: 28594403
- PMCID: PMC5520914
- DOI: 10.1038/cddis.2017.258
Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways
Abstract
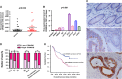
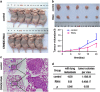
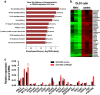
Recent studies have furthered our understanding of the function of long noncoding RNAs (lncRNAs) in numerous biological processes, including cancer. This study investigated the expression of a novel lncRNA, colorectal neoplasia differentially expressed (CRNDE), in colorectal carcinoma (CRC) tissues and cells by real-time RT-PCR and in situ hybridization, and its biological function using a series of in vitro and in vivo experiments to determine its potential as a prognostic marker and therapeutic target. CRNDE was found to be upregulated in primary CRC tissues and cells (P<0.05), and the upregulation of CRNDE expression is a powerful predictor of advanced TNM stage (P<0.05) and poor prognosis for CRC patients (P=0.002). The promoting effects of CRNDE on the cell proliferation, cell cycling and metastasis of CRC cells were confirmed both in vitro and in vivo by gain-of-function and loss-of-function experiments. Mechanistically, it was demonstrated that CRNDE could form a functional complex with heterogeneous nuclear ribonucleoprotein U-like 2 protein (hnRNPUL2) and direct the transport of hnRNPUL2 between the nucleus and cytoplasm. hnRNPUL2 that was accumulated in the cytoplasm could interact with CRNDE both physically and functionally, increasing the stability of CRNDE RNA. Moreover, gene expression profile data showed that CRNDE depletion in cells downregulated a series of genes involved in the Ras/mitogen-activated protein kinase signaling pathways. Collectively, these findings provide novel insights into the function and mechanism of lncRNA CRNDE in the pathogenesis of CRC and highlight its potential as a therapeutic target for CRC intervention.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression.Cell Death Dis. 2017 Aug 10;8(8):e2997. doi: 10.1038/cddis.2017.328. Cell Death Dis. 2017. PMID: 28796262 Free PMC article.
-
CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism.Biochim Biophys Acta. 2014 Feb;1843(2):372-86. doi: 10.1016/j.bbamcr.2013.10.016. Epub 2013 Nov 1. Biochim Biophys Acta. 2014. PMID: 24184209
-
The Long Non-Coding RNA CRNDE Promotes Colorectal Carcinoma Progression by Competitively Binding miR-217 with TCF7L2 and Enhancing the Wnt/β-Catenin Signaling Pathway.Cell Physiol Biochem. 2017;41(6):2489-2502. doi: 10.1159/000475941. Epub 2017 May 4. Cell Physiol Biochem. 2017. PMID: 28472810
-
The prognostic role of long noncoding RNA CRNDE in cancer patients: a systematic review and meta-analysis.Neoplasma. 2019 Jan 15;66(1):73-82. doi: 10.4149/neo_2018_180320N191. Epub 2018 Aug 9. Neoplasma. 2019. PMID: 30509091
-
CRNDE: A Pivotal Oncogenic Long Non-Coding RNA in Cancers.Yale J Biol Med. 2023 Dec 29;96(4):511-526. doi: 10.59249/VHYE2306. eCollection 2023 Dec. Yale J Biol Med. 2023. PMID: 38161583 Free PMC article. Review.
Cited by
-
Long non-coding RNA CRNDE promote the progression of tongue squamous cell carcinoma through regulating the PI3K/AKT/mTOR signaling pathway.RSC Adv. 2019 Jul 10;9(37):21381-21390. doi: 10.1039/c9ra01321k. eCollection 2019 Jul 5. RSC Adv. 2019. PMID: 35521355 Free PMC article.
-
Nucleolin facilitates nuclear retention of an ultraconserved region containing TRA2β4 and accelerates colon cancer cell growth.Oncotarget. 2018 Jun 1;9(42):26817-26833. doi: 10.18632/oncotarget.25510. eCollection 2018 Jun 1. Oncotarget. 2018. PMID: 29928487 Free PMC article.
-
CRNDE: An important oncogenic long non-coding RNA in human cancers.Cell Prolif. 2018 Jun;51(3):e12440. doi: 10.1111/cpr.12440. Epub 2018 Feb 5. Cell Prolif. 2018. PMID: 29405523 Free PMC article. Review.
-
Long noncoding RNAs: Undeciphered cellular codes encrypting keys of colorectal cancer pathogenesis.Cancer Lett. 2018 Mar 28;417:89-95. doi: 10.1016/j.canlet.2017.12.033. Epub 2018 Jan 3. Cancer Lett. 2018. PMID: 29306015 Free PMC article. Review.
-
LncRNA CRNDE Promotes ATG4B-Mediated Autophagy and Alleviates the Sensitivity of Sorafenib in Hepatocellular Carcinoma Cells.Front Cell Dev Biol. 2021 Aug 2;9:687524. doi: 10.3389/fcell.2021.687524. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34409031 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

