A comparison of Cox and logistic regression for use in genome-wide association studies of cohort and case-cohort design
- PMID: 28594416
- PMCID: PMC5520083
- DOI: 10.1038/ejhg.2017.78
A comparison of Cox and logistic regression for use in genome-wide association studies of cohort and case-cohort design
Abstract
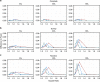
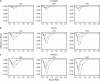
Logistic regression is often used instead of Cox regression to analyse genome-wide association studies (GWAS) of single-nucleotide polymorphisms (SNPs) and disease outcomes with cohort and case-cohort designs, as it is less computationally expensive. Although Cox and logistic regression models have been compared previously in cohort studies, this work does not completely cover the GWAS setting nor extend to the case-cohort study design. Here, we evaluated Cox and logistic regression applied to cohort and case-cohort genetic association studies using simulated data and genetic data from the EPIC-CVD study. In the cohort setting, there was a modest improvement in power to detect SNP-disease associations using Cox regression compared with logistic regression, which increased as the disease incidence increased. In contrast, logistic regression had more power than (Prentice weighted) Cox regression in the case-cohort setting. Logistic regression yielded inflated effect estimates (assuming the hazard ratio is the underlying measure of association) for both study designs, especially for SNPs with greater effect on disease. Given logistic regression is substantially more computationally efficient than Cox regression in both settings, we propose a two-step approach to GWAS in cohort and case-cohort studies. First to analyse all SNPs with logistic regression to identify associated variants below a pre-defined P-value threshold, and second to fit Cox regression (appropriately weighted in case-cohort studies) to those identified SNPs to ensure accurate estimation of association with disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Fast Algorithms for Conducting Large-Scale GWAS of Age-at-Onset Traits Using Cox Mixed-Effects Models.Genetics. 2020 May;215(1):41-58. doi: 10.1534/genetics.119.302940. Epub 2020 Mar 4. Genetics. 2020. PMID: 32132097 Free PMC article.
-
Evaluation of methodology for the analysis of 'time-to-event' data in pharmacogenomic genome-wide association studies.Pharmacogenomics. 2016 Jun;17(8):907-15. doi: 10.2217/pgs.16.19. Epub 2016 Jun 1. Pharmacogenomics. 2016. PMID: 27248145 Free PMC article.
-
Utilizing Deep Learning and Genome Wide Association Studies for Epistatic-Driven Preterm Birth Classification in African-American Women.IEEE/ACM Trans Comput Biol Bioinform. 2020 Mar-Apr;17(2):668-678. doi: 10.1109/TCBB.2018.2868667. Epub 2018 Sep 3. IEEE/ACM Trans Comput Biol Bioinform. 2020. PMID: 30183645
-
Design considerations for genetic linkage and association studies.Methods Mol Biol. 2012;850:237-62. doi: 10.1007/978-1-61779-555-8_13. Methods Mol Biol. 2012. PMID: 22307702
-
Genome-wide association studies in nephrology: using known associations for data checks.Am J Kidney Dis. 2015 Feb;65(2):217-22. doi: 10.1053/j.ajkd.2014.09.019. Epub 2014 Nov 18. Am J Kidney Dis. 2015. PMID: 25465167 Free PMC article. Review.
Cited by
-
Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients.Genome Med. 2019 Oct 9;11(1):61. doi: 10.1186/s13073-019-0672-4. Genome Med. 2019. PMID: 31597568 Free PMC article.
-
Efficient and accurate frailty model approach for genome-wide survival association analysis in large-scale biobanks.Nat Commun. 2022 Sep 16;13(1):5437. doi: 10.1038/s41467-022-32885-x. Nat Commun. 2022. PMID: 36114182 Free PMC article.
-
Thrombotic risk determined by ABO, F8, and VWF variants in a population-based cohort study.Res Pract Thromb Haemost. 2025 Apr 27;9(4):102875. doi: 10.1016/j.rpth.2025.102875. eCollection 2025 May. Res Pract Thromb Haemost. 2025. PMID: 40488174 Free PMC article.
-
Cox regression increases power to detect genotype-phenotype associations in genomic studies using the electronic health record.BMC Genomics. 2019 Nov 4;20(1):805. doi: 10.1186/s12864-019-6192-1. BMC Genomics. 2019. PMID: 31684865 Free PMC article.
-
Cox regression is robust to inaccurate EHR-extracted event time: an application to EHR-based GWAS.Bioinformatics. 2022 Apr 12;38(8):2297-2306. doi: 10.1093/bioinformatics/btac086. Bioinformatics. 2022. PMID: 35157022 Free PMC article.
References
-
- Green MS, Symons MJ: A comparison of the logistic risk function and the proportional hazards model in prospective epidemiologic studies. J Chronic Dis 1983; 36: 715–723. - PubMed
-
- Callas PW, Pastides H, Hosmer DW: Empirical comparisons of proportional hazards, Poisson, and logistic regression modeling of occupational cohort data. Am J Ind Med 1998; 33: 33–47. - PubMed
-
- Annesi I, Moreau T, Lellouch J: Efficiency of the logistic regression and Cox proportional hazards models in longitudinal studies. Stat Med 1989; 8: 1515–1521. - PubMed
Publication types
MeSH terms
Grants and funding
- RG/13/13/30194/BHF_/British Heart Foundation/United Kingdom
- G73632/MRC_/Medical Research Council/United Kingdom
- MR/L003120/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12015/1/MRC_/Medical Research Council/United Kingdom
- 268834/ERC_/European Research Council/International
- MC_UU_12015/5/MRC_/Medical Research Council/United Kingdom
- G0700463/MRC_/Medical Research Council/United Kingdom
- G66840/MRC_/Medical Research Council/United Kingdom
- RG/08/014/24067/BHF_/British Heart Foundation/United Kingdom
- SP/09/002/BHF_/British Heart Foundation/United Kingdom
- G0800270/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

