Multicopy plasmid integration in Komagataella phaffii mediated by a defective auxotrophic marker
- PMID: 28595601
- PMCID: PMC5465527
- DOI: 10.1186/s12934-017-0715-8
Multicopy plasmid integration in Komagataella phaffii mediated by a defective auxotrophic marker
Abstract
Background: A commonly used approach to improve recombinant protein production is to increase the levels of expression by providing extra-copies of a heterologous gene. In Komagataella phaffii (Pichia pastoris) this is usually accomplished by transforming cells with an expression vector carrying a drug-resistance marker following a screening for multicopy clones on plates with increasingly higher concentrations of an antibiotic. Alternatively, defective auxotrophic markers can be used for the same purpose. These markers are generally transcriptionally impaired genes lacking most of the promoter region. Among the defective markers commonly used in Saccharomyces cerevisiae is leu2-d, an allele of LEU2 which is involved in leucine metabolism. Cells transformed with this marker can recover prototrophy when they carry multiple copies of leu2-d in order to compensate the poor transcription from this defective allele.
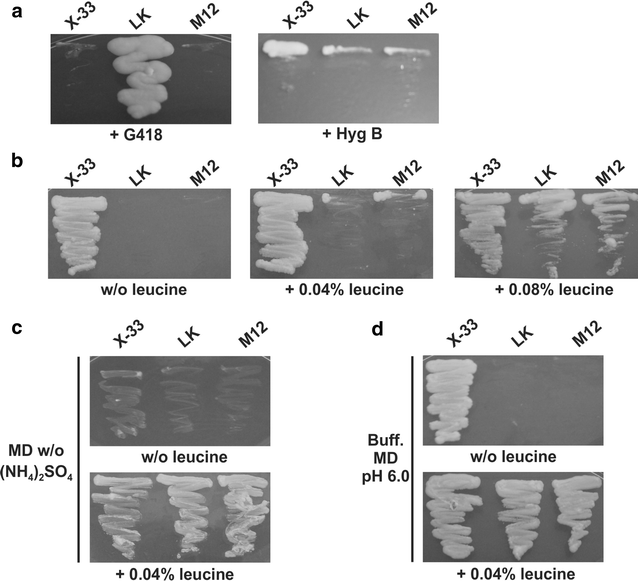
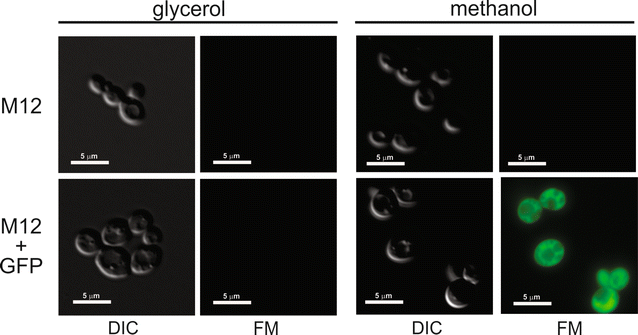
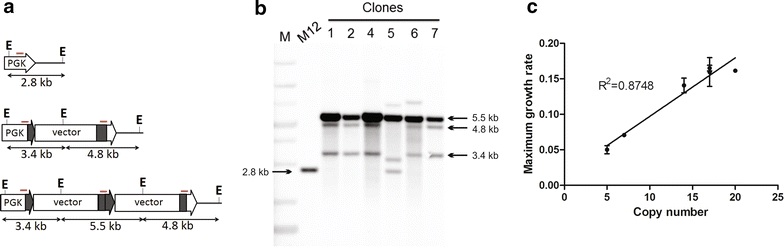
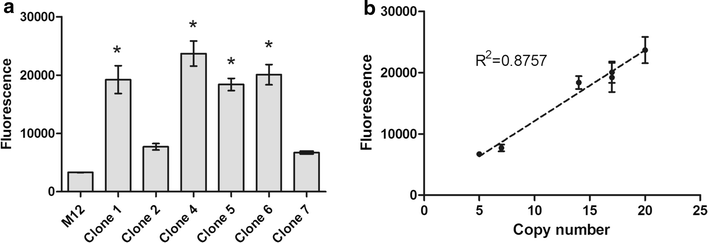
Results: A K. phaffii strain auxotrophic for leucine (M12) was constructed by disrupting endogenous LEU2. The resulting strain was successfully transformed with a vector carrying leu2-d and an EGFP (enhanced green fluorescent protein) reporter gene. Vector copy numbers were determined from selected clones which grew to different colony sizes on transformation plates. A direct correlation was observed between colony size, number of integrated vectors and EGFP production. By using this approach we were able to isolate genetically stable clones bearing as many as 20 integrated copies of the vector and with no significant effects on cell growth.
Conclusions: In this work we have successfully developed a genetic system based on a defective auxotrophic which can be applied to improve heterologous protein production in K. phaffii. The system comprises a K. phaffii leu2 strain and an expression vector carrying the defective leu2-d marker which allowed the isolation of multicopy clones after a single transformation step. Because a linear correlation was observed between copy number and heterologous protein production, this system may provide a simple approach to improve recombinant protein productivity in K. phaffii.
Keywords: Auxotrophic marker; Expression system; Komagataella phaffii; Leucine biosynthesis; Multicopy integration.
Figures


References
-
- Mellitzer A, Ruth C, Gustafsson C, Welch M, Birner-Grünberger R, Weis R, Purkarthofer T, Glieder A. Synergistic modular promoter and gene optimization to push cellulase secretion by Pichia pastoris beyond existing benchmarks. J Biotechnol. 2014;191:187–195. doi: 10.1016/j.jbiotec.2014.08.035. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

