The pepper virome: natural co-infection of diverse viruses and their quasispecies
- PMID: 28595635
- PMCID: PMC5465472
- DOI: 10.1186/s12864-017-3838-8
The pepper virome: natural co-infection of diverse viruses and their quasispecies
Abstract
Background: The co-infection of diverse viruses in a host plant is common; however, little is known about viral populations and their quasispecies in the host.
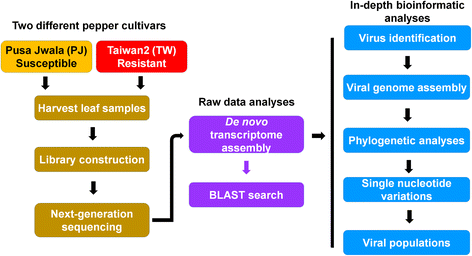
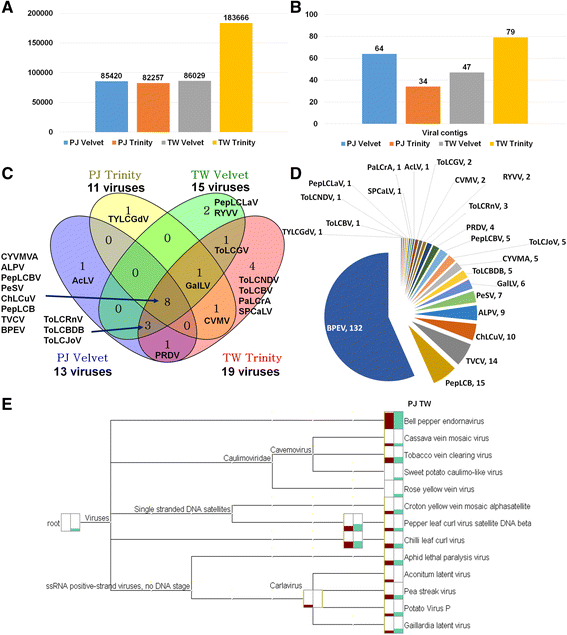
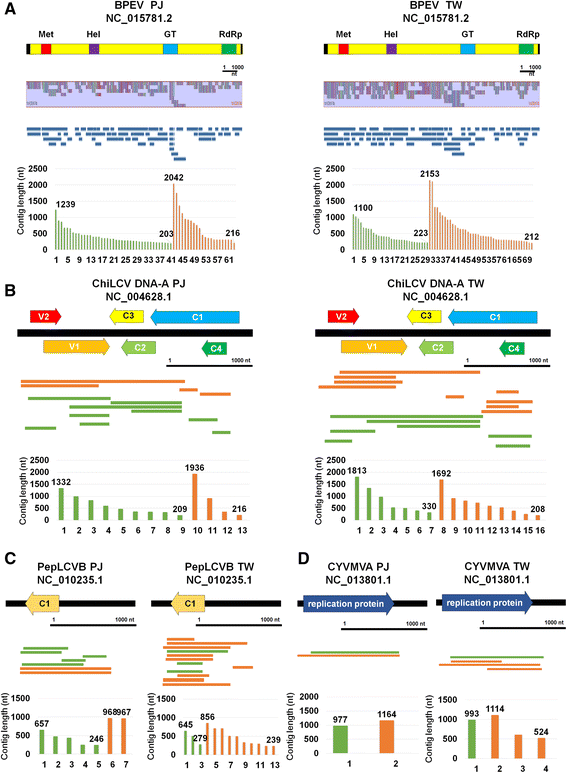
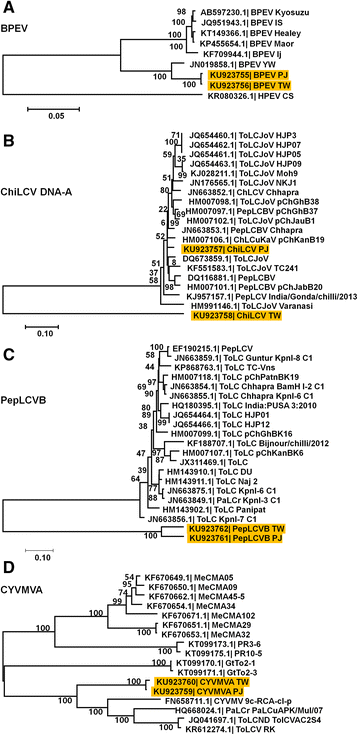
Results: Here, we report the first pepper viromes that were co-infected by different types of viral genomes. The pepper viromes are dominated by geminivirus DNA-A followed by a novel carlavirus referred to as Pepper virus A. The two pepper cultivars share similar viral populations and replications. However, the quasispecies for double-stranded RNA virus and two satellite DNAs were heterogeneous and homogenous in susceptible and resistant cultivars, respectively, indicating the quasispecies of an individual virus depends on the host.
Conclusions: Taken together, we provide the first evidence that the host plant resistant to viruses has an unrevealed antiviral system, affecting viral quasispecies, not replication.
Keywords: Cultivar; Pepper; Plant virus; Quasispecies; Virome.
Figures



Similar articles
-
Viromes of 15 Pepper (Capsicum annuum L.) Cultivars.Int J Mol Sci. 2022 Sep 10;23(18):10507. doi: 10.3390/ijms231810507. Int J Mol Sci. 2022. PMID: 36142418 Free PMC article. Review.
-
Coevolution of a Persistent Plant Virus and Its Pepper Hosts.Mol Plant Microbe Interact. 2018 Jul;31(7):766-776. doi: 10.1094/MPMI-12-17-0312-R. Epub 2018 May 30. Mol Plant Microbe Interact. 2018. PMID: 29845896
-
In silico identification of Bell pepper endornavirus from pepper transcriptomes and their phylogenetic and recombination analyses.Gene. 2016 Jan 10;575(2 Pt 3):712-7. doi: 10.1016/j.gene.2015.09.051. Epub 2015 Sep 26. Gene. 2016. PMID: 26410036
-
Complete genome sequence of a novel endornavirus isolated from hot pepper.Arch Virol. 2015 Dec;160(12):3153-6. doi: 10.1007/s00705-015-2616-7. Epub 2015 Sep 30. Arch Virol. 2015. PMID: 26424198
-
Quasispecies structure and persistence of RNA viruses.Emerg Infect Dis. 1998 Oct-Dec;4(4):521-7. doi: 10.3201/eid0404.980402. Emerg Infect Dis. 1998. PMID: 9866728 Free PMC article. Review.
Cited by
-
Robust Virome Profiling and Whole Genome Reconstruction of Viruses and Viroids Enabled by Use of Available mRNA and sRNA-Seq Datasets in Grapevine (Vitis vinifera L.).Front Microbiol. 2020 Jun 5;11:1232. doi: 10.3389/fmicb.2020.01232. eCollection 2020. Front Microbiol. 2020. PMID: 32582126 Free PMC article.
-
Enhancing Clinical Utility: Utilization of International Standards and Guidelines for Metagenomic Sequencing in Infectious Disease Diagnosis.Int J Mol Sci. 2024 Mar 15;25(6):3333. doi: 10.3390/ijms25063333. Int J Mol Sci. 2024. PMID: 38542307 Free PMC article. Review.
-
Exploring the Global Trends of Bacillus, Trichoderma and Entomopathogenic Fungi for Pathogen and Pest Control in Chili Cultivation.Saudi J Biol Sci. 2024 Aug;31(8):104046. doi: 10.1016/j.sjbs.2024.104046. Epub 2024 Jun 12. Saudi J Biol Sci. 2024. PMID: 38983130 Free PMC article. Review.
-
A Survey of Main Pepper Crop Viruses in Different Cultivation Systems for the Selection of the Most Appropriate Resistance Genes in Sensitive Local Cultivars in Northern Spain.Plants (Basel). 2022 Mar 8;11(6):719. doi: 10.3390/plants11060719. Plants (Basel). 2022. PMID: 35336600 Free PMC article.
-
Barley RNA viromes in six different geographical regions in Korea.Sci Rep. 2018 Sep 5;8(1):13237. doi: 10.1038/s41598-018-31671-4. Sci Rep. 2018. PMID: 30185900 Free PMC article.
References
-
- Alabi O, Al Rwahnih M, Jifon J, Gregg L, Crosby K, Mirkov T. First Report of Pepper vein yellows virus Infecting Pepper (Capsicum spp.) in the United States. Plant Dis. 2015;99(11):1656. https://doi.org/10.1094/PDIS-03-15-0329-PDN. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

