The effects of structure on pre-mRNA processing and stability
- PMID: 28595983
- PMCID: PMC5737760
- DOI: 10.1016/j.ymeth.2017.06.001
The effects of structure on pre-mRNA processing and stability
Abstract
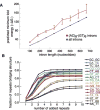
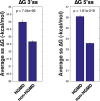
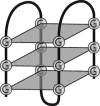
Pre-mRNA molecules can form a variety of structures, and both secondary and tertiary structures have important effects on processing, function and stability of these molecules. The prediction of RNA secondary structure is a challenging problem and various algorithms that use minimum free energy, maximum expected accuracy and comparative evolutionary based methods have been developed to predict secondary structures. However, these tools are not perfect, and this remains an active area of research. The secondary structure of pre-mRNA molecules can have an enhancing or inhibitory effect on pre-mRNA splicing. An example of enhancing structure can be found in a novel class of introns in zebrafish. About 10% of zebrafish genes contain a structured intron that forms a bridging hairpin that enforces correct splice site pairing. Negative examples of splicing include local structures around splice sites that decrease splicing efficiency and potentially cause mis-splicing leading to disease. Splicing mutations are a frequent cause of hereditary disease. The transcripts of disease genes are significantly more structured around the splice sites, and point mutations that increase the local structure often cause splicing disruptions. Post-splicing, RNA secondary structure can also affect the stability of the spliced intron and regulatory RNA interference pathway intermediates, such as pre-microRNAs. Additionally, RNA secondary structure has important roles in the innate immune defense against viruses. Finally, tertiary structure can also play a large role in pre-mRNA splicing. One example is the G-quadruplex structure, which, similar to secondary structure, can either enhance or inhibit splicing through mechanisms such as creating or obscuring RNA binding protein sites.
Keywords: Disease; ESE; ESS; G quadruplex; RNA processing; Secondary structure; Simple repeats; Splice site; Splicing; Zebrafish.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



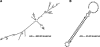
References
-
- Lorenz R, Wolfinger MT, Tanzer A, Hofacker IL. Predicting RNA secondary structures from sequence and probing data. Methods. 2016;103:86–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

