Nuclear Factor I/B: A Master Regulator of Cell Differentiation with Paradoxical Roles in Cancer
- PMID: 28596133
- PMCID: PMC5552107
- DOI: 10.1016/j.ebiom.2017.05.027
Nuclear Factor I/B: A Master Regulator of Cell Differentiation with Paradoxical Roles in Cancer
Abstract
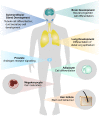
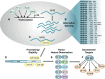
Emerging evidence indicates that nuclear factor I/B (NFIB), a transcription factor required for proper development and regulation of cellular differentiation in several tissues, also plays critical roles in cancer. Despite being a metastatic driver in small cell lung cancer and melanoma, it has become apparent that NFIB also exhibits tumour suppressive functions in many malignancies. The contradictory contributions of NFIB to both the inhibition and promotion of tumour development and progression, corroborates its diverse and context-dependent roles in many tissues and cell types. Considering the frequent involvement of NFIB in cancer, a better understanding of its multifaceted nature may ultimately benefit the development of novel strategies for the management of a broad spectrum of malignancies. Here we discuss recent findings which bring to light NFIB as a crucial and paradoxical player in cancer.
Keywords: Cancer; Cellular differentiation; Developmental transcription factor; NFIB; Oncogene; Tumour suppressor.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
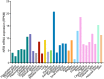

References
-
- Andreasen S., Persson M., Kiss K., Homoe P., Heegaard S., Stenman G. Genomic profiling of a combined large cell neuroendocrine carcinoma of the submandibular gland. Oncol. Rep. 2016;35:2177–2182. - PubMed
-
- Arlt M.F., Durkin S.G., Ragland R.L., Glover T.W. Common fragile sites as targets for chromosome rearrangements. DNA Repair. 2006;5:1126–1135. - PubMed
-
- Becker-Santos D.D., Thu K.L., English J.C., Pikor L.A., Martinez V.D., Zhang M., Vucic E.A., Luk M.T., Carraro A., Korbelik J. Developmental transcription factor NFIB is a putative target of oncofetal miRNAs and is associated with tumour aggressiveness in lung adenocarcinoma. J. Pathol. 2016;240:161–172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

