Yeast genetic interaction screen of human genes associated with amyotrophic lateral sclerosis: identification of MAP2K5 kinase as a potential drug target
- PMID: 28596290
- PMCID: PMC5580709
- DOI: 10.1101/gr.211649.116
Yeast genetic interaction screen of human genes associated with amyotrophic lateral sclerosis: identification of MAP2K5 kinase as a potential drug target
Abstract
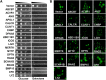
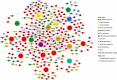
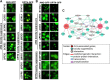
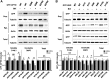
To understand disease mechanisms, a large-scale analysis of human-yeast genetic interactions was performed. Of 1305 human disease genes assayed, 20 genes exhibited strong toxicity in yeast. Human-yeast genetic interactions were identified by en masse transformation of the human disease genes into a pool of 4653 homozygous diploid yeast deletion mutants with unique barcode sequences, followed by multiplexed barcode sequencing to identify yeast toxicity modifiers. Subsequent network analyses focusing on amyotrophic lateral sclerosis (ALS)-associated genes, such as optineurin (OPTN) and angiogenin (ANG), showed that the human orthologs of the yeast toxicity modifiers of these ALS genes are enriched for several biological processes, such as cell death, lipid metabolism, and molecular transport. When yeast genetic interaction partners held in common between human OPTN and ANG were validated in mammalian cells and zebrafish, MAP2K5 kinase emerged as a potential drug target for ALS therapy. The toxicity modifiers identified in this study may deepen our understanding of the pathogenic mechanisms of ALS and other devastating diseases.
© 2017 Jo et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


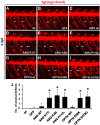
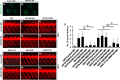
References
-
- Anborgh PH, Godin C, Pampillo M, Dhami GK, Dale LB, Cregan SP, Truant R, Ferguson SS. 2005. Inhibition of metabotropic glutamate receptor signaling by the huntingtin-binding protein optineurin. J Biol Chem 280: 34840–34848. - PubMed
-
- Babin PJ, Goizet C, Raldua D. 2014. Zebrafish models of human motor neuron diseases: advantages and limitations. Prog Neurobiol 118C: 36–58. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
