Clear Cell Renal Cell Carcinoma is linked to Epithelial-to-Mesenchymal Transition and to Fibrosis
- PMID: 28596300
- PMCID: PMC5471444
- DOI: 10.14814/phy2.13305
Clear Cell Renal Cell Carcinoma is linked to Epithelial-to-Mesenchymal Transition and to Fibrosis
Erratum in
-
Corrigendum.Physiol Rep. 2018 Apr;6(8):e13671. doi: 10.14814/phy2.13671. Physiol Rep. 2018. PMID: 29687608 Free PMC article. No abstract available.
Abstract
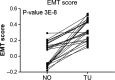
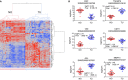
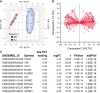
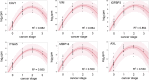
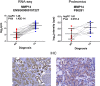
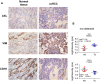
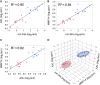
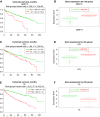
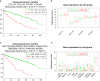
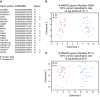
Clear cell renal cell carcinoma (ccRCC) represents the most common type of kidney cancer with high mortality in its advanced stages. Our study aim was to explore the correlation between tumor epithelial-to-mesenchymal transition (EMT) and patient survival. Renal biopsies of tumorous and adjacent nontumorous tissue were taken with a 16 g needle from our patients (n = 26) undergoing partial or radical nephrectomy due to ccRCC RNA sequencing libraries were generated using Illumina TruSeq® Access library preparation protocol and TruSeq Small RNA library preparation kit. Next generation sequencing (NGS) was performed on Illumina HiSeq2500. Comparative analysis of matched sample pairs was done using the Bioconductor Limma/voom R-package. Liquid chromatography-tandem mass spectrometry and immunohistochemistry were applied to measure and visualize protein abundance. We detected an increased generic EMT transcript score in ccRCC Gene expression analysis showed augmented abundance of AXL and MMP14, as well as down-regulated expression of KL (klotho). Moreover, microRNA analyses demonstrated a positive expression correlation of miR-34a and its targets MMP14 and AXL Survival analysis based on a subset of genes from our list EMT-related genes in a publicly available dataset showed that the EMT genes correlated with ccRCC patient survival. Several of these genes also play a known role in fibrosis. Accordingly, recently published classifiers of solid organ fibrosis correctly identified EMT-affected tumor samples and were correlated with patient survival. EMT in ccRCC linked to fibrosis is associated with worse survival and may represent a target for novel therapeutic interventions.
Keywords: Clear cell renal cell carcinoma; epithelial‐to‐mesenchymal transition; fibrosis.
© 2017 The Authors. Physiological Reports published by Wiley Periodicals, Inc. on behalf of The Physiological Society and the American Physiological Society.
Figures
Similar articles
-
Development and confirmation of potential gene classifiers of human clear cell renal cell carcinoma using next-generation RNA sequencing.Scand J Urol. 2016 Dec;50(6):452-462. doi: 10.1080/21681805.2016.1238007. Epub 2016 Oct 14. Scand J Urol. 2016. PMID: 27739342
-
Transcriptome Sequencing (RNAseq) Enables Utilization of Formalin-Fixed, Paraffin-Embedded Biopsies with Clear Cell Renal Cell Carcinoma for Exploration of Disease Biology and Biomarker Development.PLoS One. 2016 Feb 22;11(2):e0149743. doi: 10.1371/journal.pone.0149743. eCollection 2016. PLoS One. 2016. PMID: 26901863 Free PMC article.
-
The Axl-Regulating Tumor Suppressor miR-34a Is Increased in ccRCC but Does Not Correlate with Axl mRNA or Axl Protein Levels.PLoS One. 2015 Aug 19;10(8):e0135991. doi: 10.1371/journal.pone.0135991. eCollection 2015. PLoS One. 2015. PMID: 26287733 Free PMC article.
-
A lncRNA TCL6-miR-155 Interaction Regulates the Src-Akt-EMT Network to Mediate Kidney Cancer Progression and Metastasis.Cancer Res. 2021 Mar 15;81(6):1500-1512. doi: 10.1158/0008-5472.CAN-20-0832. Epub 2021 Jan 26. Cancer Res. 2021. PMID: 33500248 Free PMC article.
-
circPTCH1 promotes invasion and metastasis in renal cell carcinoma via regulating miR-485-5p/MMP14 axis.Theranostics. 2020 Aug 29;10(23):10791-10807. doi: 10.7150/thno.47239. eCollection 2020. Theranostics. 2020. PMID: 32929380 Free PMC article.
Cited by
-
Integrating bioinformatic analysis and detailed experiments reveal an EMT-related biomarker for clear cell renal cell carcinoma.Cancer Med. 2023 Sep;12(18):19320-19336. doi: 10.1002/cam4.6504. Epub 2023 Sep 7. Cancer Med. 2023. PMID: 37676078 Free PMC article.
-
Transgelin Contributes to a Poor Response of Metastatic Renal Cell Carcinoma to Sunitinib Treatment.Biomedicines. 2021 Sep 3;9(9):1145. doi: 10.3390/biomedicines9091145. Biomedicines. 2021. PMID: 34572331 Free PMC article.
-
Obesity-Dependent Adipokine Chemerin Suppresses Fatty Acid Oxidation to Confer Ferroptosis Resistance.Cancer Discov. 2021 Aug;11(8):2072-2093. doi: 10.1158/2159-8290.CD-20-1453. Epub 2021 Mar 23. Cancer Discov. 2021. PMID: 33757970 Free PMC article.
-
LncRNAs in the Regulation of Genes and Signaling Pathways through miRNA-Mediated and Other Mechanisms in Clear Cell Renal Cell Carcinoma.Int J Mol Sci. 2021 Oct 17;22(20):11193. doi: 10.3390/ijms222011193. Int J Mol Sci. 2021. PMID: 34681854 Free PMC article. Review.
-
Primary renal sarcomas: imaging features and discrimination from non-sarcoma renal tumors.Eur Radiol. 2022 Feb;32(2):981-989. doi: 10.1007/s00330-021-08201-4. Epub 2021 Jul 31. Eur Radiol. 2022. PMID: 34331576 Free PMC article.
References
-
- Baker, A. H. , Edwards D. R., and Murphy G.. 2002. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J. Cell Sci. 115:3719–3727. - PubMed
-
- Chen, D. , Gassenmaier M., Maruschke M., Riesenberg R., Pohla H., Stief C. G., et al. 2014. Expression and prognostic significance of a comprehensive epithelial‐mesenchymal transition gene set in renal cell carcinoma. J. Urol. 191:479–486. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

