Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells
- PMID: 28596481
- PMCID: PMC5564523
- DOI: 10.18632/oncotarget.18108
Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells
Abstract
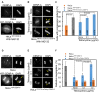
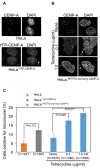
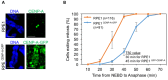
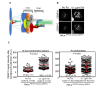
Chromosomal instability (CIN) is a hallmark of many cancers and a major contributor to tumorigenesis. Centromere and kinetochore associated proteins such as the evolutionarily conserved centromeric histone H3 variant CENP-A, associate with centromeric DNA for centromere function and chromosomal stability. Stringent regulation of cellular CENP-A levels prevents its mislocalization in yeast and flies to maintain genome stability. CENP-A overexpression and mislocalization are observed in several cancers and reported to be associated with increased invasiveness and poor prognosis. We examined whether there is a direct relationship between mislocalization of overexpressed CENP-A and CIN using HeLa and chromosomally stable diploid RPE1 cell lines as model systems. Our results show that mislocalization of overexpressed CENP-A to chromosome arms leads to chromosome congression defects, lagging chromosomes, micronuclei formation and a delay in mitotic exit. CENP-A overexpressing cells showed altered localization of centromere and kinetochore associated proteins such as CENP-C, CENP-T and Nuf2 leading to weakened native kinetochores as shown by reduced interkinetochore distance and CIN. Importantly, our results show that mislocalization of CENP-A to chromosome arms is one of the major contributors for CIN as depletion of histone chaperone DAXX prevents CENP-A mislocalization and rescues the reduced interkinetochore distance and CIN phenotype in CENP-A overexpressing cells. In summary, our results establish that CENP-A overexpression and mislocalization result in a CIN phenotype in human cells. This study provides insights into how overexpression of CENP-A may contribute to CIN in cancers and underscore the importance of understanding the pathways that prevent CENP-A mislocalization for genome stability.
Keywords: CENP-A; Chromosome Section; DAXX; cancer; centromeres; chromosomal instability.
Conflict of interest statement
No potential conflict of interest
Figures



Similar articles
-
Protein Phosphatase 1 Regulatory Subunit PNUTS Prevents CENP-A Mislocalization and Chromosomal Instability.Mol Cell Biol. 2025;45(5):185-197. doi: 10.1080/10985549.2025.2487010. Epub 2025 Apr 24. Mol Cell Biol. 2025. PMID: 40270285
-
The histone H3/H4 chaperone CHAF1B prevents the mislocalization of CENP-A for chromosomal stability.J Cell Sci. 2023 May 15;136(10):jcs260944. doi: 10.1242/jcs.260944. Epub 2023 May 31. J Cell Sci. 2023. PMID: 37129573 Free PMC article.
-
Reduced gene dosage of histone H4 prevents CENP-A mislocalization and chromosomal instability in Saccharomyces cerevisiae.Genetics. 2021 May 17;218(1):iyab033. doi: 10.1093/genetics/iyab033. Genetics. 2021. PMID: 33751052 Free PMC article.
-
Recent insights into mechanisms preventing ectopic centromere formation.Open Biol. 2021 Sep;11(9):210189. doi: 10.1098/rsob.210189. Epub 2021 Sep 8. Open Biol. 2021. PMID: 34493071 Free PMC article. Review.
-
Guarding the Genome: CENP-A-Chromatin in Health and Cancer.Genes (Basel). 2020 Jul 16;11(7):810. doi: 10.3390/genes11070810. Genes (Basel). 2020. PMID: 32708729 Free PMC article. Review.
Cited by
-
Oncogenic lncRNAs alter epigenetic memory at a fragile chromosomal site in human cancer cells.Sci Adv. 2022 Mar 4;8(9):eabl5621. doi: 10.1126/sciadv.abl5621. Epub 2022 Mar 2. Sci Adv. 2022. PMID: 35235361 Free PMC article.
-
Histone variants and chromatin structure, update of advances.Comput Struct Biotechnol J. 2022 Dec 5;21:299-311. doi: 10.1016/j.csbj.2022.12.002. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36582440 Free PMC article. Review.
-
Clinical Implications of Chromosomal Instability (CIN) and Kinetochore Abnormalities in Breast Cancers.Mol Diagn Ther. 2019 Dec;23(6):707-721. doi: 10.1007/s40291-019-00420-2. Mol Diagn Ther. 2019. PMID: 31372940 Review.
-
Emerging Single-Cell Technological Approaches to Investigate Chromatin Dynamics and Centromere Regulation in Human Health and Disease.Int J Mol Sci. 2021 Aug 16;22(16):8809. doi: 10.3390/ijms22168809. Int J Mol Sci. 2021. PMID: 34445507 Free PMC article. Review.
-
Ubiquitin-dependent proteolysis of KNL2 driven by APC/CCDC20 is critical for centromere integrity and mitotic fidelity.Plant Cell. 2025 Jul 1;37(7):koaf164. doi: 10.1093/plcell/koaf164. Plant Cell. 2025. PMID: 40561118 Free PMC article.
References
-
- Weaver BA, Cleveland DW. Does Anauploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–67. 0.1016/j.ceb.2006.10.002. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

