Transcriptome sequencing of Tessaratoma papillosa antennae to identify and analyze expression patterns of putative olfaction genes
- PMID: 28596537
- PMCID: PMC5465196
- DOI: 10.1038/s41598-017-03306-7
Transcriptome sequencing of Tessaratoma papillosa antennae to identify and analyze expression patterns of putative olfaction genes
Abstract
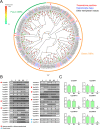
Studies on insect olfaction have increased our understanding of insect's chemosensory system and chemical ecology, and have improved pest control strategies based on insect behavior. In this study, we assembled the antennal transcriptomes of the lychee giant stink bug, Tessaratoma papillosa, by using next generation sequencing to identify the major olfaction gene families in this species. In total, 59 odorant receptors, 14 ionotropic receptors (8 antennal IRs), and 33 odorant binding proteins (28 classic OBPs and 5 plus-C OBPs) were identified from the male and female antennal transcriptomes. Analyses of tissue expression profiles revealed that all 59 OR transcripts, 2 of the 8 antennal IRs, and 6 of the 33 OBPs were primarily expressed in the antennae, suggesting their putative role in olfaction. The sex-biased expression patterns of these antenna-predominant genes suggested that they may have important functions in the reproductive behavior of these insects. This is the first report that provides a comprehensive resource to future studies on olfaction in the lychee giant stink bug.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Steinbrecht RA. Pore structures in insect olfactory sensilla: A review of data and concepts. Int. J. Insect Morphol. 1997;26:229–245. doi: 10.1016/S0020-7322(97)00024-X. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

