The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis
- PMID: 28596563
- PMCID: PMC5465203
- DOI: 10.1038/s41598-017-03321-8
The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis
Abstract
The SEPT9 gene methylation assay is the first FDA-approved blood assay for colorectal cancer (CRC) screening. Fecal immunochemical test (FIT), FIT-DNA test and CEA assay are also in vitro diagnostic (IVD) tests used in CRC screening. This meta-analysis aims to review the SEPT9 assay performance and compare it with other IVD CRC screening tests. By searching the Ovid MEDLINE, EMBASE, CBMdisc and CJFD database, 25 out of 180 studies were identified to report the SEPT9 assay performance. 2613 CRC cases and 6030 controls were included, and sensitivity and specificity were used to evaluate its performance at various algorithms. 1/3 algorithm exhibited the best sensitivity while 2/3 and 1/1 algorithm exhibited the best balance between sensitivity and specificity. The performance of the blood SEPT9 assay is superior to that of the serum protein markers and the FIT test in symptomatic population, while appeared to be less potent than FIT and FIT-DNA tests in asymptomatic population. In conclusion, 1/3 algorithm is recommended for CRC screening, and 2/3 or 1/1 algorithms are suitable for early detection for diagnostic purpose. The SEPT9 assay exhibited better performance in symptomatic population than in asymptomatic population.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
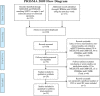

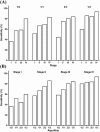
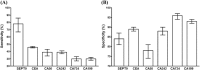
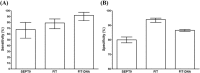


References
-
- National Cancer Institute. Survival rate for colorectal cancer by stage, Physician Data Query, Treatment, Health Professionals 1999 (1999).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

