In-depth Profiling of MvfR-Regulated Small Molecules in Pseudomonas aeruginosa after Quorum Sensing Inhibitor Treatment
- PMID: 28596760
- PMCID: PMC5442231
- DOI: 10.3389/fmicb.2017.00924
In-depth Profiling of MvfR-Regulated Small Molecules in Pseudomonas aeruginosa after Quorum Sensing Inhibitor Treatment
Abstract
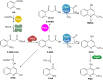
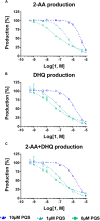
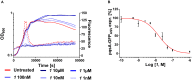
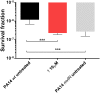
Pseudomonas aeruginosa is a Gram-negative bacterium, which causes opportunistic infections in immuno-compromised individuals. Due to its multiple resistances toward antibiotics, the development of new drugs is required. Interfering with Quorum Sensing (QS), a cell-to-cell communication system, has shown to be highly efficient in reducing P. aeruginosa pathogenicity. One of its QS systems employs Pseudomonas Quinolone Signal (PQS) and 4-hydroxy-2-heptylquinoline (HHQ) as signal molecules. Both activate the transcriptional regulator MvfR (Multiple Virulence Factor Regulator), also called PqsR, driving the production of QS molecules as well as toxins and biofilm formation. The aim of this work was to elucidate the effects of QS inhibitors (QSIs), such as MvfR antagonists and PqsBC inhibitors, on the biosynthesis of the MvfR-regulated small molecules 2'-aminoacetophenone (2-AA), dihydroxyquinoline (DHQ), HHQ, PQS, and 4-hydroxy-2-heptylquinoline-N-oxide (HQNO). The employed synthetic MvfR antagonist fully inhibited pqs small molecule formation showing expected sigmoidal dose-response curves for 2-AA, HQNO, HHQ and PQS. Surprisingly, DHQ levels were enhanced at lower antagonist concentrations followed by a full suppression at higher QSI amounts. This particular bi-phasic profile hinted at the accumulation of a biosynthetic intermediate resulting in the observed overproduction of the shunt product DHQ. Additionally, investigations on PqsBC inhibitors showed a reduction of MvfR natural ligands, while increased 2-AA, DHQ and HQNO levels compared to the untreated cells were detected. Moreover, PqsBC inhibitors did not show any significant effect in PA14 pqsC mutant demonstrating their target selectivity. As 2-AA is important for antibacterial tolerance, the QSIs were evaluated in their capability to attenuate persistence. Indeed, persister cells were reduced along with 2-AA inhibition resulting from MvfR antagonism, but not from PqsBC inhibition. In conclusion, antagonizing MvfR using a dosage capable of fully suppressing this QS system will lead to a favorable therapeutic outcome as DHQ overproduction is avoided and bacterial persistence is reduced.
Keywords: 2′-aminoacetophenone; MvfR; Pseudomonas aeruginosa; Quorum Sensing Inhibitors; dihydroxyquinoline; persistence; quinolones.
Figures





References
-
- Drees S. L., Li C., Prasetya F., Saleem M., Dreveny I., Williams P., et al. (2016). PqsBC, a condensing enzyme in the biosynthesis of the Pseudomonas aeruginosa quinolone signal: crystal structure, inhibition, and reaction mechanism. J. Biol. Chem. 291 6610–6624. 10.1074/jbc.M115.708453 - DOI - PMC - PubMed
-
- Dulcey C. E., Dekimpe V., Fauvelle D.-A., Milot S., Groleau M.-C., Doucet N., et al. (2013). The end of an old hypothesis: the Pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem. Biol. 20 1481–1491. 10.1016/j.chembiol.2013.09.021 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

