A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses
- PMID: 28598328
- PMCID: PMC5511008
- DOI: 10.7554/eLife.26437
A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses
Abstract
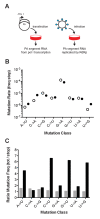
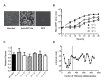
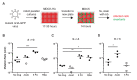
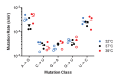
Influenza virus' low replicative fidelity contributes to its capacity for rapid evolution. Clonal sequencing and fluctuation tests have suggested that the influenza virus mutation rate is 2.7 × 10-6 - 3.0 × 10-5 substitutions per nucleotide per strand copied (s/n/r). However, sequencing assays are biased toward mutations with minimal fitness impacts and fluctuation tests typically investigate only a subset of all possible single nucleotide mutations. We developed a fluctuation test based on reversion to fluorescence in a set of virally encoded mutant green fluorescent proteins, which allowed us to measure the rates of selectively neutral mutations representative of the twelve different mutation types. We measured an overall mutation rate of 1.8 × 10-4 s/n/r for PR8 (H1N1) and 2.5 × 10-4 s/n/r for Hong Kong 2014 (H3N2) and a transitional bias of 2.7-3.6. Our data suggest that each replicated genome will have an average of 2-3 mutations and highlight the importance of mutational load in influenza virus evolution.
Keywords: diversity; evolution; evolutionary biology; genomics; infectious disease; microbiology; mutation rate; virus.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

Comment in
-
A new twist in measuring mutation rates.Elife. 2017 Jul 14;6:e29586. doi: 10.7554/eLife.29586. Elife. 2017. PMID: 28708485 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

