Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition
- PMID: 28598437
- PMCID: PMC5472775
- DOI: 10.1038/ncomms15775
Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition
Abstract
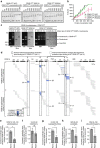
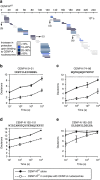
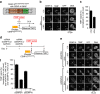
Maintaining centromere identity relies upon the persistence of the epigenetic mark provided by the histone H3 variant, centromere protein A (CENP-A), but the molecular mechanisms that underlie its remarkable stability remain unclear. Here, we define the contributions of each of the three candidate CENP-A nucleosome-binding domains (two on CENP-C and one on CENP-N) to CENP-A stability using gene replacement and rapid protein degradation. Surprisingly, the most conserved domain, the CENP-C motif, is dispensable. Instead, the stability is conferred by the unfolded central domain of CENP-C and the folded N-terminal domain of CENP-N that becomes rigidified 1,000-fold upon crossbridging CENP-A and its adjacent nucleosomal DNA. Disrupting the 'arginine anchor' on CENP-C for the nucleosomal acidic patch disrupts the CENP-A nucleosome structural transition and removes CENP-A nucleosomes from centromeres. CENP-A nucleosome retention at centromeres requires a core centromeric nucleosome complex where CENP-C clamps down a stable nucleosome conformation and CENP-N fastens CENP-A to the DNA.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Similar articles
-
Decoding the centromeric nucleosome through CENP-N.Elife. 2017 Dec 27;6:e33442. doi: 10.7554/eLife.33442. Elife. 2017. PMID: 29280735 Free PMC article.
-
Molecular basis of CENP-C association with the CENP-A nucleosome at yeast centromeres.Genes Dev. 2017 Oct 1;31(19):1958-1972. doi: 10.1101/gad.304782.117. Epub 2017 Oct 26. Genes Dev. 2017. PMID: 29074736 Free PMC article.
-
Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere.Science. 2015 May 8;348(6235):699-703. doi: 10.1126/science.1259308. Science. 2015. PMID: 25954010 Free PMC article.
-
Orchestrating the Specific Assembly of Centromeric Nucleosomes.Prog Mol Subcell Biol. 2017;56:165-192. doi: 10.1007/978-3-319-58592-5_7. Prog Mol Subcell Biol. 2017. PMID: 28840237 Free PMC article. Review.
-
Structure of the CENP-A nucleosome and its implications for centromeric chromatin architecture.Genes Genet Syst. 2011;86(6):357-64. doi: 10.1266/ggs.86.357. Genes Genet Syst. 2011. PMID: 22451475 Review.
Cited by
-
Diverse mechanisms of centromere specification.Curr Biol. 2021 Nov 22;31(22):R1491-R1504. doi: 10.1016/j.cub.2021.09.083. Curr Biol. 2021. PMID: 34813757 Free PMC article. Review.
-
Mobility of kinetochore proteins measured by FRAP analysis in living cells.Chromosome Res. 2022 Mar;30(1):43-57. doi: 10.1007/s10577-021-09678-x. Epub 2022 Jan 8. Chromosome Res. 2022. PMID: 34997387 Free PMC article.
-
Dynamic cell cycle-dependent phosphorylation modulates CENP-L-CENP-N centromere recruitment.Mol Biol Cell. 2022 Sep 1;33(10):ar87. doi: 10.1091/mbc.E22-06-0239. Epub 2022 Jul 13. Mol Biol Cell. 2022. PMID: 35830614 Free PMC article.
-
High-resolution analysis of human centromeric chromatin.Life Sci Alliance. 2025 Jan 23;8(4):e202402819. doi: 10.26508/lsa.202402819. Print 2025 Apr. Life Sci Alliance. 2025. PMID: 39848706 Free PMC article.
-
The Mis6 inner kinetochore subcomplex maintains CENP-A nucleosomes against centromeric non-coding transcription during mitosis.Commun Biol. 2022 Aug 15;5(1):818. doi: 10.1038/s42003-022-03786-y. Commun Biol. 2022. PMID: 35970865 Free PMC article.
References
-
- Earnshaw W. C. & Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–321 (1985). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

