Joint modeling of genetically correlated diseases and functional annotations increases accuracy of polygenic risk prediction
- PMID: 28598966
- PMCID: PMC5482506
- DOI: 10.1371/journal.pgen.1006836
Joint modeling of genetically correlated diseases and functional annotations increases accuracy of polygenic risk prediction
Abstract
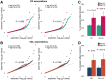
Accurate prediction of disease risk based on genetic factors is an important goal in human genetics research and precision medicine. Advanced prediction models will lead to more effective disease prevention and treatment strategies. Despite the identification of thousands of disease-associated genetic variants through genome-wide association studies (GWAS) in the past decade, accuracy of genetic risk prediction remains moderate for most diseases, which is largely due to the challenges in both identifying all the functionally relevant variants and accurately estimating their effect sizes. In this work, we introduce PleioPred, a principled framework that leverages pleiotropy and functional annotations in genetic risk prediction for complex diseases. PleioPred uses GWAS summary statistics as its input, and jointly models multiple genetically correlated diseases and a variety of external information including linkage disequilibrium and diverse functional annotations to increase the accuracy of risk prediction. Through comprehensive simulations and real data analyses on Crohn's disease, celiac disease and type-II diabetes, we demonstrate that our approach can substantially increase the accuracy of polygenic risk prediction and risk population stratification, i.e. PleioPred can significantly better separate type-II diabetes patients with early and late onset ages, illustrating its potential clinical application. Furthermore, we show that the increment in prediction accuracy is significantly correlated with the genetic correlation between the predicted and jointly modeled diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Chatterjee N, Shi J, Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;advance online publication. http://www.nature.com/nrg/journal/vaop/ncurrent/abs/nrg.2016.27.html-sup.... - PMC - PubMed
-
- Li C, Yang C, Gelernter J, Zhao H. Improving genetic risk prediction by leveraging pleiotropy. Human genetics. 2014;133(5):639–50. doi: 10.1007/s00439-013-1401-5 - DOI - PMC - PubMed
-
- Maier R, Moser G, Chen G-B, Ripke S, Coryell W, Potash JB, et al. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. The American Journal of Human Genetics. 2015;96(2):283–94. doi: 10.1016/j.ajhg.2014.12.006 - DOI - PMC - PubMed
-
- Minnier J, Yuan M, Liu JS, Cai T. Risk classification with an adaptive naive bayes kernel machine model. Journal of the American Statistical Association. 2015;110(509):393–404. doi: 10.1080/01621459.2014.908778 - DOI - PMC - PubMed
-
- Speed D, Balding DJ. MultiBLUP: improved SNP-based prediction for complex traits. Genome research. 2014;24(9):1550–7. doi: 10.1101/gr.169375.113 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

