Selection of reference genes for quantitative real-time RT-PCR assays in different morphological forms of dimorphic zygomycetous fungus Benjaminiella poitrasii
- PMID: 28598997
- PMCID: PMC5466344
- DOI: 10.1371/journal.pone.0179454
Selection of reference genes for quantitative real-time RT-PCR assays in different morphological forms of dimorphic zygomycetous fungus Benjaminiella poitrasii
Abstract
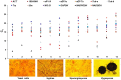
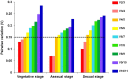
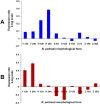
Benjaminiella poitrasii, a dimorphic non-pathogenic zygomycetous fungus, exhibits a morphological yeast (Y) to hypha (H) reversible transition in the vegetative phase, sporangiospores (S) in the asexual phase and zygospores (Z) in the sexual phase. To study the gene expression across these diverse morphological forms, suitable reference genes are required. In the present study, 13 genes viz. ACT, 18S rRNA, eEF1α, eEF-Tu,eIF-1A, Tub-α, Tub-b, Ubc, GAPDH, Try, WS-21, NADGDH and NADPGDH were evaluated for their potential as a reference, particularly for studying gene expression during the Y-H reversible transition and also for other asexual and sexual life stages of B. poitrasii. Analysis of RT-qPCR data using geNorm, normFinder and BestKeeper software revealed that genes such as Ubc, 18S rRNA and WS-21 were expressed at constant levels in each given subset of RNA samples from all the morphological phases of B. poitrasii. Therefore, these reference genes can be used to elucidate the role of morpho-genes in B. poitrasii. Further, use of the two most stably expressed genes (Ubc and WS-21) to normalize the expression of the ornithine decarboxylase gene (Bpodc) in different morphological forms of B. poitrasii, generated more reliable results, indicating that our selection of reference genes was appropriate.
Conflict of interest statement
Figures


Similar articles
-
Morphology-associated expression of NADP-dependent glutamate dehydrogenases during yeast-mycelium transition of a dimorphic fungus Benjaminiella poitrasii.Antonie Van Leeuwenhoek. 2004 May;85(4):327-34. doi: 10.1023/B:ANTO.0000020384.26238.48. Antonie Van Leeuwenhoek. 2004. PMID: 15031645
-
Molecular studies of NAD- and NADP-glutamate dehydrogenases decipher the conundrum of yeast-hypha dimorphism in zygomycete Benjaminiella poitrasii.FEMS Yeast Res. 2019 Dec 1;19(8):foz074. doi: 10.1093/femsyr/foz074. FEMS Yeast Res. 2019. PMID: 31644791
-
Significance of NADP/NAD glutamate dehydrogenase ratio in the dimorphic behavior of Benjaminiella poitrasii and its morphological mutants.J Bacteriol. 1992 Jun;174(11):3723-8. doi: 10.1128/jb.174.11.3723-3728.1992. J Bacteriol. 1992. PMID: 1592824 Free PMC article.
-
Possible involvement of cyclic adenosine 3',5'-monophosphate in the regulation of NADP-/NAD-glutamate dehydrogenase ratio and in yeast-mycelium transition of Benjaminiella poitrasii.J Bacteriol. 1993 Sep;175(18):6052-5. doi: 10.1128/jb.175.18.6052-6055.1993. J Bacteriol. 1993. PMID: 8397189 Free PMC article.
-
The zygomycetous fungus, Benjaminiella poitrasii contains a large family of differentially regulated chitin synthase genes.Fungal Genet Biol. 2002 Aug;36(3):215-23. doi: 10.1016/s1087-1845(02)00015-4. Fungal Genet Biol. 2002. PMID: 12135577
Cited by
-
Mucor circinelloides: Growth, Maintenance, and Genetic Manipulation.Curr Protoc Microbiol. 2018 May;49(1):e53. doi: 10.1002/cpmc.53. Epub 2018 Apr 27. Curr Protoc Microbiol. 2018. PMID: 30040216 Free PMC article.
-
Reference genes for gene expression analysis in the fungal pathogen Neonectria ditissima and their use demonstrating expression up-regulation of candidate virulence genes.PLoS One. 2020 Nov 13;15(11):e0238157. doi: 10.1371/journal.pone.0238157. eCollection 2020. PLoS One. 2020. PMID: 33186359 Free PMC article.
-
The Sporothrix schenckii Gene Encoding for the Ribosomal Protein L6 Has Constitutive and Stable Expression and Works as an Endogenous Control in Gene Expression Analysis.Front Microbiol. 2017 Sep 1;8:1676. doi: 10.3389/fmicb.2017.01676. eCollection 2017. Front Microbiol. 2017. PMID: 28919888 Free PMC article.
-
Expression Patterns in Reductive Iron Assimilation and Functional Consequences during Phagocytosis of Lichtheimia corymbifera, an Emerging Cause of Mucormycosis.J Fungi (Basel). 2021 Apr 3;7(4):272. doi: 10.3390/jof7040272. J Fungi (Basel). 2021. PMID: 33916756 Free PMC article.
-
Selection and validation of reference genes for qRT-PCR analysis of gene expression in Microsporum canis growing under different adhesion-inducing conditions.Sci Rep. 2018 Jan 19;8(1):1197. doi: 10.1038/s41598-018-19680-9. Sci Rep. 2018. PMID: 29352152 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

