Identification of candidate drugs for the treatment of metastatic osteosarcoma through a subpathway analysis method
- PMID: 28599440
- PMCID: PMC5453166
- DOI: 10.3892/ol.2017.5953
Identification of candidate drugs for the treatment of metastatic osteosarcoma through a subpathway analysis method
Abstract
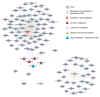
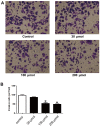
Osteosarcoma (OS) is the third most frequent type of cancer in adolescents and represents >56% of all bone tumors. In addition, metastatic OS frequently demonstrates resistance to conventional chemotherapy; thus, the development of novel therapeutic agents for the treatment of patients with metastatic OS is warranted. In the present study, the metabolic mechanisms underlying OS metastasis were investigated using a subpathway analysis method and lead to the identification of candidate drugs for the treatment of metastatic OS. Using the GSE14827 microarray dataset from the Gene Expression Omnibus database, 546 differentially expressed genes were identified between samples from patients with OS who did or did not develop metastatic OS. Furthermore, nine significantly enriched metabolic subpathways were identified, which may be involved in OS metastasis. Finally, using an integrated analysis of metastatic OS-associated subpathways and drug-affected subpathways, 98 small molecule drug candidates capable of targeting the metastatic OS-associated subpathways were identified. This method identified existing anti-cancer drugs, including semustine, in addition to predicting potential drugs, such as lansoprazole, for the treatment of metastatic OS. Transwell and wound healing assays demonstrated that lansoprazole reduced the invasiveness and migration of U2OS cells. These small molecule drug candidates identified through a bioinformatics approach may provide insights into novel therapy options for the treatment of patients with metastatic OS.
Keywords: drugs; metabolic subpathways; metastatic osteosarcoma; network.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
