Laboratory Evolution of a Biotin-Requiring Saccharomyces cerevisiae Strain for Full Biotin Prototrophy and Identification of Causal Mutations
- PMID: 28600311
- PMCID: PMC5541218
- DOI: 10.1128/AEM.00892-17
Laboratory Evolution of a Biotin-Requiring Saccharomyces cerevisiae Strain for Full Biotin Prototrophy and Identification of Causal Mutations
Abstract
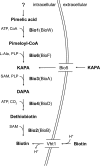
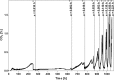
Biotin prototrophy is a rare, incompletely understood, and industrially relevant characteristic of Saccharomyces cerevisiae strains. The genome of the haploid laboratory strain CEN.PK113-7D contains a full complement of biotin biosynthesis genes, but its growth in biotin-free synthetic medium is extremely slow (specific growth rate [μ] ≈ 0.01 h-1). Four independent evolution experiments in repeated batch cultures and accelerostats yielded strains whose growth rates (μ ≤ 0.36 h-1) in biotin-free and biotin-supplemented media were similar. Whole-genome resequencing of these evolved strains revealed up to 40-fold amplification of BIO1, which encodes pimeloyl-coenzyme A (CoA) synthetase. The additional copies of BIO1 were found on different chromosomes, and its amplification coincided with substantial chromosomal rearrangements. A key role of this gene amplification was confirmed by overexpression of BIO1 in strain CEN.PK113-7D, which enabled growth in biotin-free medium (μ = 0.15 h-1). Mutations in the membrane transporter genes TPO1 and/or PDR12 were found in several of the evolved strains. Deletion of TPO1 and PDR12 in a BIO1-overexpressing strain increased its specific growth rate to 0.25 h-1 The effects of null mutations in these genes, which have not been previously associated with biotin metabolism, were nonadditive. This study demonstrates that S. cerevisiae strains that carry the basic genetic information for biotin synthesis can be evolved for full biotin prototrophy and identifies new targets for engineering biotin prototrophy into laboratory and industrial strains of this yeast.IMPORTANCE Although biotin (vitamin H) plays essential roles in all organisms, not all organisms can synthesize this vitamin. Many strains of baker's yeast, an important microorganism in industrial biotechnology, contain at least some of the genes required for biotin synthesis. However, most of these strains cannot synthesize biotin at all or do so at rates that are insufficient to sustain fast growth and product formation. Consequently, this expensive vitamin is routinely added to baker's yeast cultures. In this study, laboratory evolution in biotin-free growth medium yielded new strains that grew as fast in the absence of biotin as in its presence. By analyzing the DNA sequences of evolved biotin-independent strains, mutations were identified that contributed to this ability. This work demonstrates full biotin independence of an industrially relevant yeast and identifies mutations whose introduction into other yeast strains may reduce or eliminate their biotin requirements.
Keywords: Saccharomyces cerevisiae; adaptive laboratory evolution; biotin; prototrophy; reverse metabolic engineering; vitamin biosynthesis; whole-genome sequencing.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- Gailiusis J, Rinne RW, Benedict C. 1964. Pyruvate- oxaloacetate exchange reaction in baker's yeast. Biochim Biophys Acta 92:595–601. - PubMed
-
- Losada M, Canovas J, Ruiz A. 1964. Oxaloacetate, citramalate and glutamate formation from pyruvate in baker's yeast. Biochem Z 340:60–74. - PubMed
-
- Sumrada RA, Cooper TG. 1982. Urea carboxylase and allophanate hydrolase are components of a multifunctional protein in yeast. J Biol Chem 257:9119–9127. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

