The Oxidative Stress Response in Caenorhabditis elegans Requires the GATA Transcription Factor ELT-3 and SKN-1/Nrf2
- PMID: 28600327
- PMCID: PMC5560797
- DOI: 10.1534/genetics.116.198788
The Oxidative Stress Response in Caenorhabditis elegans Requires the GATA Transcription Factor ELT-3 and SKN-1/Nrf2
Abstract
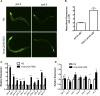
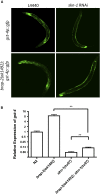
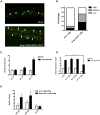
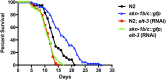
Cellular damage caused by reactive oxygen species is believed to be a major contributor to age-associated diseases. Previously, we characterized the Caenorhabditis elegans Brap2 ortholog (BRAP-2) and found that it is required to prevent larval arrest in response to elevated levels of oxidative stress. Here, we report that C. elegans brap-2 mutants display increased expression of SKN-1-dependent, phase II detoxification enzymes that is dependent on PMK-1 (a p38 MAPK C. elegans ortholog). An RNA-interference screen was conducted using a transcription factor library to identify genes required for increased expression of the SKN-1 target gst-4 in brap-2 mutants. We identified ELT-3, a member of the GATA transcription factor family, as a positive regulator of gst-4p::gfp expression. We found that ELT-3 interacts with SKN-1 to activate gst-4 transcription in vitro and that elt-3 is required for enhanced gst-4 expression in the brap-2(ok1492) mutant in vivo Furthermore, nematodes overexpressing SKN-1 required ELT-3 for life-span extension. Taken together, these results suggest a model where BRAP-2 acts as negative regulator of SKN-1 through inhibition of p38 MAPK activity, and that the GATA transcription factor ELT-3 is required along with SKN-1 for the phase II detoxification response in C. elegans.
Keywords: BRAP-2; ELT-3; GATA transcription factors; SKN-1; oxidative stress.
Copyright © 2017 by the Genetics Society of America.
Figures



Similar articles
-
BRAP-2 promotes DNA damage induced germline apoptosis in C. elegans through the regulation of SKN-1 and AKT-1.Cell Death Differ. 2018 Jul;25(7):1276-1288. doi: 10.1038/s41418-017-0038-7. Epub 2018 Jan 22. Cell Death Differ. 2018. PMID: 29358669 Free PMC article.
-
The Caenorhabditis elegans Oxidative Stress Response Requires the NHR-49 Transcription Factor.G3 (Bethesda). 2018 Dec 10;8(12):3857-3863. doi: 10.1534/g3.118.200727. G3 (Bethesda). 2018. PMID: 30297383 Free PMC article.
-
The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult C. elegans.PLoS Genet. 2015 May 27;11(5):e1005265. doi: 10.1371/journal.pgen.1005265. eCollection 2015 May. PLoS Genet. 2015. PMID: 26016853 Free PMC article.
-
The Caenorhabditis elegans intestine.Wiley Interdiscip Rev Dev Biol. 2013 May-Jun;2(3):347-67. doi: 10.1002/wdev.93. Epub 2012 Oct 9. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 23799580 Review.
-
SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans.Free Radic Biol Med. 2015 Nov;88(Pt B):290-301. doi: 10.1016/j.freeradbiomed.2015.06.008. Epub 2015 Aug 5. Free Radic Biol Med. 2015. PMID: 26232625 Free PMC article. Review.
Cited by
-
Nematicidal Characterization of Solanum nigrum and Mentha arvensis Leaf Extracts Using Caenorhabditis elegans as a Model Organism.ACS Omega. 2023 Feb 16;8(10):9454-9463. doi: 10.1021/acsomega.2c08124. eCollection 2023 Mar 14. ACS Omega. 2023. PMID: 36936282 Free PMC article.
-
A Damage Sensor Associated with the Cuticle Coordinates Three Core Environmental Stress Responses in Caenorhabditis elegans.Genetics. 2018 Apr;208(4):1467-1482. doi: 10.1534/genetics.118.300827. Epub 2018 Feb 27. Genetics. 2018. PMID: 29487136 Free PMC article.
-
Acute, reproductive, and developmental toxicity of essential oils assessed with alternative in vitro and in vivo systems.Arch Toxicol. 2021 Feb;95(2):673-691. doi: 10.1007/s00204-020-02945-6. Epub 2020 Nov 7. Arch Toxicol. 2021. PMID: 33159585 Free PMC article.
-
Immunity-linked genes are stimulated by a membrane stress pathway linked to Golgi function and the ARF-1 GTPase.Sci Adv. 2023 Dec 8;9(49):eadi5545. doi: 10.1126/sciadv.adi5545. Epub 2023 Dec 6. Sci Adv. 2023. PMID: 38055815 Free PMC article.
-
Transcriptomic analysis reveals molecular mechanism by which Chinese olive fruit prolongs lifespan of Caenorhabditis elegans.NPJ Sci Food. 2025 May 30;9(1):90. doi: 10.1038/s41538-025-00456-1. NPJ Sci Food. 2025. PMID: 40447648 Free PMC article.
References
-
- Bishop N. A., Guarente L., 2007. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447: 545–549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

