Direct competition between DNA binding factors highlights the role of Krüppel-like Factor 1 in the erythroid/megakaryocyte switch
- PMID: 28600522
- PMCID: PMC5466599
- DOI: 10.1038/s41598-017-03289-5
Direct competition between DNA binding factors highlights the role of Krüppel-like Factor 1 in the erythroid/megakaryocyte switch
Abstract
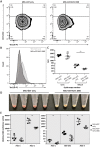
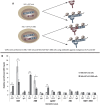
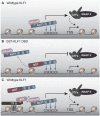
The Krüppel-like factor (KLF) family of transcription factors play critical roles in haematopoiesis. KLF1, the founding member of the family, has been implicated in the control of both erythropoiesis and megakaryopoiesis. Here we describe a novel system using an artificial dominant negative isoform of KLF1 to investigate the role of KLF1 in the erythroid/megakaryocytic switch in vivo. We developed murine cell lines stably overexpressing a GST-KLF1 DNA binding domain fusion protein (GST-KLF1 DBD), as well as lines expressing GST only as a control. Interestingly, overexpression of GST-KLF1 DBD led to an overall reduction in erythroid features and an increase in megakaryocytic features indicative of a reduced function of endogenous KLF1. We simultaneously compared in vivo DNA occupancy of both endogenous KLF1 and GST-KLF1 DBD by ChIP qPCR. Here we found that GST-KLF1 DBD physically displaces endogenous KLF1 at a number of loci, providing novel in vivo evidence of direct competition between DNA binding proteins. These results highlight the role of KLF1 in the erythroid/megakaryocyte switch and suggest that direct competition between transcription factors with similar consensus sequences is an important mechanism in transcriptional regulation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

