Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration
- PMID: 28600982
- PMCID: PMC5466586
- DOI: 10.1016/j.redox.2017.05.015
Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration
Abstract
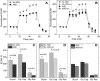
Age-related macular degeneration (AMD) is the leading cause of blindness among older adults. It has been suggested that mitochondrial defects in the retinal pigment epithelium (RPE) underlies AMD pathology. To test this idea, we developed primary cultures of RPE to ask whether RPE from donors with AMD differ in their metabolic profile compared with healthy age-matched donors. Analysis of gene expression, protein content, and RPE function showed that these cultured cells replicated many of the cardinal features of RPE in vivo. Using the Seahorse Extracellular Flux Analyzer to measure bioenergetics, we observed RPE from donors with AMD exhibited reduced mitochondrial and glycolytic function compared with healthy donors. RPE from AMD donors were also more resistant to oxidative inactivation of these two energy-producing pathways and were less susceptible to oxidation-induced cell death compared with cells from healthy donors. Investigation of the potential mechanism responsible for differences in bioenergetics and resistance to oxidative stress showed RPE from AMD donors had increased PGC1α protein as well as differential expression of multiple genes in response to an oxidative challenge. Based on our data, we propose that cultured RPE from donors phenotyped for the presence or absence of AMD provides an excellent model system for studying "AMD in a dish". Our results are consistent with the ideas that (i) a bioenergetics crisis in the RPE contributes to AMD pathology, and (ii) the diseased environment in vivo causes changes in the cellular profile that are retained in vitro.
Keywords: 6 max) Age-related macular degeneration; Antioxidants; Glycolytic function; Mitochondrial function; Oxidative stress; Retinal pigment epithelium.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures





References
-
- Alliance for Eye and Vision Research, 20th Anniversary Special Report. October. 〈http://www.eyeresearch.org/pdf/ValueBrochure/Value_of_Vision_Research_B..., 2013.
-
- Blenkinsop T.A., Salero E., Stern J.H., Temple S. The culture and maintenance of functional retinal pigment epithelial monolayers from adult human eyes. Methods Mol. Biol. 2016;945:45–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

