A Fast and Accurate Algorithm to Test for Binary Phenotypes and Its Application to PheWAS
- PMID: 28602423
- PMCID: PMC5501775
- DOI: 10.1016/j.ajhg.2017.05.014
A Fast and Accurate Algorithm to Test for Binary Phenotypes and Its Application to PheWAS
Abstract
The availability of electronic health record (EHR)-based phenotypes allows for genome-wide association analyses in thousands of traits and has great potential to enable identification of genetic variants associated with clinical phenotypes. We can interpret the phenome-wide association study (PheWAS) result for a single genetic variant by observing its association across a landscape of phenotypes. Because a PheWAS can test thousands of binary phenotypes, and most of them have unbalanced or often extremely unbalanced case-control ratios (1:10 or 1:600, respectively), existing methods cannot provide an accurate and scalable way to test for associations. Here, we propose a computationally fast score-test-based method that estimates the distribution of the test statistic by using the saddlepoint approximation. Our method is much (∼100 times) faster than the state-of-the-art Firth's test. It can also adjust for covariates and control type I error rates even when the case-control ratio is extremely unbalanced. Through application to PheWAS data from the Michigan Genomics Initiative, we show that the proposed method can control type I error rates while replicating previously known association signals even for traits with a very small number of cases and a large number of controls.
Keywords: GWAS; PheWAS; rare variants; saddlepoint approximation; single-variant test; unbalanced case-control.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

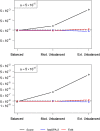

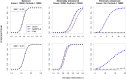

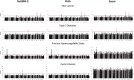
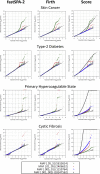
References
-
- Denny J.C., Crawford D.C., Ritchie M.D., Bielinski S.J., Basford M.A., Bradford Y., Chai H.S., Bastarache L., Zuvich R., Peissig P. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am. J. Hum. Genet. 2011;89:529–542. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

