Engineering biological systems using automated biofoundries
- PMID: 28602523
- PMCID: PMC5544601
- DOI: 10.1016/j.ymben.2017.06.003
Engineering biological systems using automated biofoundries
Abstract
Engineered biological systems such as genetic circuits and microbial cell factories have promised to solve many challenges in the modern society. However, the artisanal processes of research and development are slow, expensive, and inconsistent, representing a major obstacle in biotechnology and bioengineering. In recent years, biological foundries or biofoundries have been developed to automate design-build-test engineering cycles in an effort to accelerate these processes. This review summarizes the enabling technologies for such biofoundries as well as their early successes and remaining challenges.
Copyright © 2017 International Metabolic Engineering Society. Published by Elsevier Inc. All rights reserved.
Figures


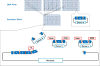

References
-
- Appleton E, Tao JH, Haddock T, Densmore D. Interactive assembly algorithms for molecular cloning. Nat Methods. 2014;11:657–+. - PubMed
-
- Araki M, Cox RS, Makiguchi H, Ogawa T, Taniguchi T, Miyaoku K, Nakatsui M, Hara KY, Kondo A. M-path: A compass for navigating potential metabolic pathways. Bioinformatics. 2015;31:905–911. - PubMed
-
- Bao ZH, Xiao H, Lang J, Zhang L, Xiong X, Sun N, Si T, Zhao HM. Homology-Integrated CRISPR-Cas (HI-CRISPR) System for One-Step Multigene Disruption in Saccharomyces cerevisiae. ACS Synth Biol. 2015;4:585–594. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

